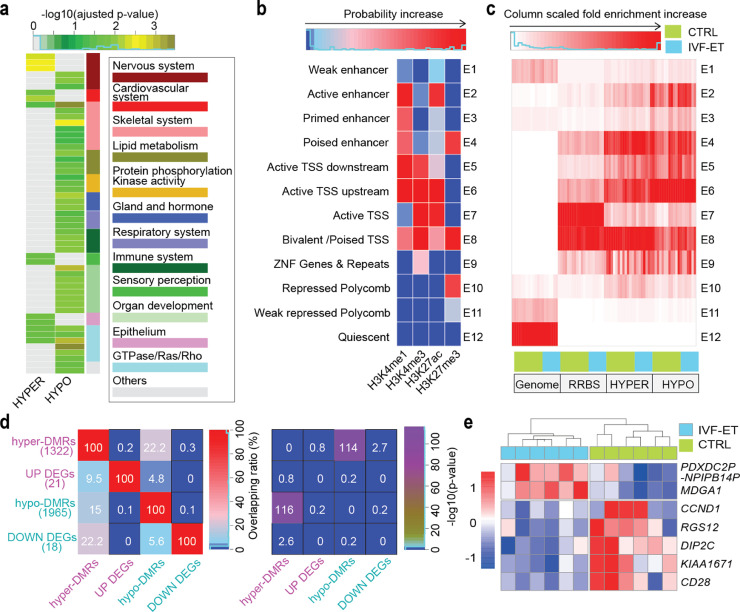
Fig. 3.
IVF effects on the epigenome of the offspring. (a) Gene ontology (biological process) analysis for the hyper- and hypo-DMRs of IVF-ET versus CTRL, all enrichment terms with adjusted p-value < 0.05 were grouped and shown. The p-value was determined by hypergeometric test and adjusted by BH method. (b) Based on the four histone modifications in all neonatal samples, 12 chromatin states were defined by chromHMM. Each row of the heatmap corresponds to a specific chromatin state, and each column corresponds to a different histone mark, H3K4me1, H3K4me3, H3K27me3, or H3K27ac. (c) In each neonatal sample of CTRL and IVF-ET groups, column scaled fold enrichment of the chromatin states at genome-wide regions (Genome), RRBS covered regions (RRBS), hyper- and hypo-DMRs (HYPER and HYPO) was shown. (d) Heatmap showed overlapping ratio among associated genes of DMRs and DEGs for IVF-ET versus CTRL (left), and their statistical significances (right) determined by hypergeometric test. (e) Heatmap showed the expression level of DEGs overlapped with the associated genes of DMRs (PDXDC2P-NPIPB14P, MDGA1, CCND1, RGS12, DIP2C, KIAA1671, and CD28) among neonatal samples in CTRL and IVF-ET.