Abstract
Background
Among the paraspinal muscles, the structure and function of the lumbar multifidus (LM) has become of great interest to researchers and clinicians involved in lower back pain and muscle rehabilitation. Ultrasound (US) imaging of the LM muscle is a useful clinical tool which can be used in the assessment of muscle morphology and function. US is widely used due to its portability, cost-effectiveness, and ease-of-use. In order to assess muscle function, quantitative information of the LM must be extracted from the US image by means of manual segmentation. However, manual segmentation requires a higher level of training and experience and is characterized by a level of difficulty and subjectivity associated with image interpretation. Thus, the development of automated segmentation methods is warranted and would strongly benefit clinicians and researchers. The aim of this study is to provide a database which will contribute to the development of automated segmentation algorithms of the LM.
Construction and content
This database provides the US ground truth of the left and right LM muscles at the L5 level (in prone and standing positions) of 109 young athletic adults involved in Concordia University’s varsity teams. The LUMINOUS database contains the US images with their corresponding manually segmented binary masks, serving as the ground truth. The purpose of the database is to enable development and validation of deep learning algorithms used for automatic segmentation tasks related to the assessment of the LM cross-sectional area (CSA) and echo intensity (EI). The LUMINOUS database is publicly available at http://data.sonography.ai.
Conclusion
The development of automated segmentation algorithms based on this database will promote the standardization of LM measurements and facilitate comparison among studies. Moreover, it can accelerate the clinical implementation of quantitative muscle assessment in clinical and research settings.
Keywords: Ultrasound imaging, Paraspinal muscle, Lumbar multifidus muscle, Segmentation
Background
The paraspinal muscles (e.g. multifidus and erector spinae muscles) are a group of three muscles that originate from the occipital bone and continue down the spine to the sacrum [1]. Among the lumbar muscles, biomechanical studies have provided evidence for the importance of the lumbar multifidus muscle (LM) and its role in the dynamic stabilization and segmental control of the lumbar spine [2]. Over two thirds of the stiffness of the spine is attributed to the behaviour of the multifidi, establishing the LM’s importance in the neutral zone [3]. The neutral zone is described the range of intervertebral motion where spinal movement can occur with minimal internal resistance from the spine [4, 5]. As opposed to all the lumbar muscles, the LM has the characteristic of being a large multifascicular muscle that has a high cross-sectional area (CSA) [2, 4, 6]. As such, its structure allows for large forces to be generated over smaller ranges of operation [4]. This further supports the LM’s role of being a unit dedicated to providing lumbar spine stability [4, 7]. Therefore, the LM’s morphology (e.g. size, composition, asymmetry) and function (e.g. contractile ability) have become of great interest to researchers and clinicians involved in lower back pain (LBP) and muscle rehabilitation [2].
LBP is one of the most prevalent medical complaints, and it is estimated that between 60% to 80% of the population will experience at least one episode in their lifetime [8–10]. More importantly, the recurrence rate is extremely high and this common musculoskeletal condition is very disabling, and it severely affects the quality of life. Furthermore, it is projected to have an even higher personal and socio-economic burden as the world’s population ages [11, 12]. A large body of evidence confirmed that LM muscle structural changes (e.g. atrophy and increased in fatty infiltration) and functional deficits (e.g. decreased or increased contraction) occur in patients with LBP [13–16]. Along with LM and spinal dysfunction, such changes are also associated with lower physical function [17–20], poorer surgical outcomes [21, 22], and the recurrence of LBP symptoms [23, 24].
To date, magnetic resonance imaging (MRI), computed tomography (CT) scan, and ultrasound (US) have been used to quantify paraspinal muscle morphology. While MRI provides excellent soft tissue contrast and resolution and is the gold-standard imaging modality, it remains costly and its accessibility is limited. US is a portable, cost-effective, and non-ionizing imaging modality, providing a non-invasive method to obtain real-time in-vivo images for the assessment of LM morphology and function [25]. More specifically, US has been used to quantify the LM CSA, and CSA side-to-side asymmetry, as well as LM thickness in resting and contracted states to assess muscle activation (e.g. contraction) [26–28]. Additionally, measurements of the echo intensity (EI) can also be obtained using computer-aided gray scale analysis. EI has been investigated in studies related to muscle morphology, changes related to neuromuscular disorders, and studies investigating the relationship between muscle EI and size [29, 30]. Moreover, EI is used as an indicator of fatty infiltration and connective tissue which can be subsequently used to assess muscle quality [30–32].
Biomechanical modelling of the spine requires accurate measurements of the LM CSA for use in analytical processes that determine levels of LM wasting or injury [33]. In US, CSA measurements can be obtained by imaging the transverse section of LM [2]. The muscle’s border is then delineated from the rest of the surrounding tissue through manual segmentation. US examination requires training and experience, and the analysis and interpretation of the images are prone to subjectivity. Additionally, US assessments in the clinical setting are subject to issues concerning procedural and measurement reliability [25]. Procedural and measurement reliability are defined as the ability of an examiner to consistently and repeatedly perform the imaging procedure and measurements of the region of interest in the muscle, respectively [25]. However, due to the shape of the LM varying from one patient to another, and from one spinal level to another, examiners performing manual segmentations often encounter technical challenges affecting the quality and reliability of these measures. One of the major limitations of LM segmentation in US images is to determine the boundaries between the LM and the surrounding tissues [34]. Thus, the manual segmentation process of US images is highly rater-dependent, error prone, and can be labour intensive, which can limit its clinical applicability [35]. Therefore, the development of automated segmentation methods is warranted and would strongly benefit clinicians and researchers by decreasing the workload while simultaneously producing accurate and reliable segmentations that are comparable to expert manual segmentations [36].
The advent of deep learning has introduced many tools which are currently used to carry out various diagnostic tasks in medical US analysis. Moreover, as deep learning is being widely used in medical US analysis, its application continues to benefit from the ongoing research efforts made to further its state-of-the-art performance [36, 37]. Although recent efforts and studies have emphasized on US segmentation tasks using deep learning approaches, there is a limited amount of literature pertaining to the segmentation of skeletal muscle [37, 38]. Thus, it would be beneficial to support US segmentation tasks of musculoskeletal muscles such as the LM. Nevertheless, the development of automated segmentation methods requires manually annotated clinical datasets, which are currently scarce. Therefore, the purpose of this work is to provide a publicly available US database with the ground truth of the left and right LM at the L5 level, in both prone and standing positions, intended for the development of automated segmentation algorithms. To the best of our knowledge, this is the first publicly available US database of LM muscle.
Construction and content
Subjects’ description
The database contains 109 US datasets of young athletic adult volunteers who are involved in select varsity teams at Concordia University (64 males, 45 females, age: 21.1 ±1.7). The participants identified themselves among to the following choices for ethnical backgrounds: Black, White, Hispanic, and Other.
Subjects’ characteristics
Subjects’ characteristics (sex, age [years], ethnicity, weight [kg], body mass index (BMI) [kg/m2], CSA [cm2], and mean EI) are listed in Table 1.
Table 1.
Subjects’ characteristics and measurements of the right/left LM at the L5 level when subjects were either in the prone or standing positions
| Right MF Prone: | Left MF Prone: | Right MF Standing: | Left MF Standing: | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Sex | Age | Ethnicity | Weight | Height | BMI | CSA | Mean | frame | CSA | Mean | frame | CSA | Mean | frame | CSA | Mean | frame |
| [years] | [kg] | [m] | [kg/m2] | [cm2] | EI | # | [cm2] | EI | # | [cm2] | EI | # | [cm2] | EI | # | |||
| 1 | M | 22 | White | 84 | 1.85 | 24.6 | 11.46 | 68.34 | 1 | 13.05 | 69.46 | 3 | 9.82 | 63.74 | 23 | 11.80 | 60.60 | 27 |
| 2 | F | 22 | White | 89 | 1.63 | 24.1 | 12.89 | 94.54 | 1 | 13.17 | 98.18 | 4 | 10.24 | 85.47 | 32 | 9.65 | 84.04 | 37 |
| 3 | M | 19 | Black | 73 | 1.78 | 23.0 | 6.95 | 33.06 | 1 | 7.25 | 33.68 | 1 | 8.75 | 40.19 | 22 | 7.98 | 23.24 | 22 |
| 4 | F | 21 | White | 88 | 1.62 | 27.3 | 8.89 | 63.14 | 1 | 8.72 | 59.69 | 1 | 9.21 | 64.11 | 29 | 9.31 | 54.35 | 29 |
| 5 | M | 23 | White | 86 | 1.78 | 28.2 | 8.68 | 86.29 | 1 | 8.19 | 84.63 | 1 | 7.40 | 65.77 | 28 | 8.80 | 66.18 | 28 |
| 6 | F | 22 | White | 95 | 1.86 | 25.6 | 7.79 | 67.50 | 1 | 6.96 | 59.28 | 5 | 9.98 | 57.78 | 28 | 9.81 | 46.34 | 32 |
| 7 | F | 23 | White | 64 | 1.62 | 24.1 | 7.46 | 44.32 | 2 | 6.40 | 44.74 | 4 | 10.01 | 54.16 | 24 | 10.14 | 57.95 | 26 |
| 8 | M | 21 | White | 87 | 1.77 | 27.5 | 10.68 | 48.21 | 1 | 9.68 | 44.67 | 5 | 12.34 | 27.43 | 29 | ex | ex | ex |
| 9 | F | 25 | Black | 76 | 1.80 | 23.5 | 10.56 | 70.40 | 1 | 9.94 | 82.77 | 3 | 10.57 | 53.37 | 26 | 11.27 | 61.31 | 31 |
| 10 | M | 21 | White | 88 | 1.87 | 27.1 | 9.89 | 52.15 | 1 | 11.17 | 54.94 | 4 | 11.50 | 37.47 | 33 | 10.70 | 48.93 | 34 |
| 11 | M | 20 | White | 82 | 1.88 | 21.9 | 8.26 | 79.01 | 1 | 7.48 | 72.55 | 3 | ex | ex | ex | ex | ex | ex |
| 12 | F | 23 | White | 89 | 1.73 | 23.4 | 10.23 | 72.46 | 1 | 11.69 | 74.34 | 4 | ex | ex | ex | ex | ex | ex |
| 13 | M | 20 | White | 81 | 1.85 | 24.9 | 13.17 | 64.68 | 1 | 13.43 | 58.56 | 5 | ex | ex | ex | 11.25 | 44.91 | 32 |
| 14 | M | 19 | White | 70 | 1.60 | 25.5 | 8.58 | 48.64 | 1 | 8.48 | 43.82 | 3 | 9.57 | 49.65 | 28 | 8.82 | 44.76 | 34 |
| 15 | M | 18 | White | 76 | 1.91 | 22.5 | 10.00 | 50.48 | 1 | 9.79 | 61.31 | 3 | 11.21 | 69.93 | 18 | 11.20 | 61.93 | 19 |
| 16 | F | 22 | White | 85 | 1.61 | 25.1 | 8.25 | 79.87 | 11 | 9.44 | 83.99 | 12 | 7.10 | 81.21 | 36 | 7.28 | 62.27 | 37 |
| 17 | M | 20 | Black | 83 | 1.77 | 26.6 | 9.21 | 31.17 | 1 | 9.25 | 43.04 | 5 | 11.29 | 39.09 | 33 | 10.40 | 34.36 | 37 |
| 18 | M | 22 | White | 87 | 1.74 | 24.8 | 8.50 | 50.98 | 1 | 8.77 | 53.05 | 5 | 9.09 | 53.48 | 23 | 9.61 | 52.55 | 31 |
| 19 | F | 23 | White | 76 | 1.65 | 24.6 | 10.35 | 85.04 | 1 | 10.30 | 82.09 | 4 | ex | ex | ex | ex | ex | ex |
| 20 | F | 21 | White | 71 | 1.94 | 26.3 | 8.49 | 77.54 | 1 | 7.37 | 72.83 | 1 | 9.64 | 63.46 | 22 | 9.25 | 52.45 | 22 |
| 21 | M | 21 | White | 87 | 1.68 | 26.9 | 9.25 | 47.25 | 1 | 10.16 | 52.29 | 6 | 10.81 | 49.18 | 27 | 10.82 | 46.62 | 32 |
| 22 | F | 22 | White | 99 | 1.69 | 25.0 | 6.66 | 70.99 | 5 | 7.18 | 77.96 | 5 | 8.91 | 84.42 | 22 | 8.59 | 79.93 | 22 |
| 23 | M | 20 | White | 81 | 1.72 | 27.3 | 12.66 | 41.62 | 1 | 11.79 | 39.47 | 4 | 10.40 | 51.32 | 29 | 10.83 | 44.37 | 30 |
| 24 | M | 21 | Other | 93 | 1.84 | 27.4 | 12.17 | 64.33 | 3 | 11.83 | 71.12 | 3 | 12.02 | 72.10 | 20 | 11.04 | 65.10 | 20 |
| 25 | F | 20 | White | 64 | 1.64 | 23.7 | 8.94 | 44.57 | 1 | 8.14 | 51.17 | 6 | ex | ex | ex | 9.14 | 39.66 | 39 |
| 26 | M | 19 | White | 95 | 1.80 | 29.3 | 9.74 | 44.58 | 1 | 10.02 | 38.41 | 4 | ex | ex | ex | ex | ex | ex |
| 27 | F | 19 | White | 67 | 1.62 | 25.6 | 6.50 | 40.24 | 1 | 6.73 | 47.87 | 1 | 9.06 | 50.93 | 24 | 7.71 | 42.21 | 24 |
| 28 | M | 22 | White | 90 | 1.90 | 25.0 | 13.10 | 52.43 | 3 | 13.47 | 53.25 | 5 | 13.91 | 53.12 | 30 | ex | ex | ex |
| 29 | M | 21 | Asian | 87 | 1.69 | 25.0 | 10.24 | 68.14 | 1 | 9.19 | 64.25 | 6 | 9.61 | 34.43 | 32 | 9.86 | 42.92 | 36 |
| 30 | M | 21 | Black | 85 | 1.78 | 27.0 | 15.50 | 40.57 | 1 | 14.06 | 32.05 | 3 | 15.67 | 27.29 | 25 | 15.93 | 30.84 | 26 |
| 31 | M | 22 | White | 85 | 1.89 | 25.1 | 10.90 | 55.20 | 1 | 12.08 | 48.72 | 4 | 11.94 | 33.51 | 27 | 11.55 | 36.26 | 30 |
| 32 | F | 21 | White | 86 | 1.63 | 22.8 | 9.94 | 79.33 | 1 | 9.22 | 70.38 | 6 | 11.72 | 65.04 | 31 | 10.95 | 61.74 | 32 |
| 33 | F | 22 | White | 86 | 1.64 | 28.1 | 10.36 | 63.54 | 1 | 9.32 | 73.73 | 1 | 9.91 | 66.21 | 22 | 8.95 | 54.39 | 22 |
| 34 | F | 23 | White | 85 | 1.68 | 22.0 | 7.86 | 67.51 | 1 | 7.06 | 56.19 | 3 | 8.74 | 50.89 | 24 | 9.14 | 43.63 | 28 |
| 35 | F | 19 | Black | 72 | 1.78 | 21.0 | 12.10 | 51.58 | 1 | 11.63 | 49.40 | 4 | 12.07 | 54.51 | 30 | 12.29 | 50.08 | 34 |
| 36 | F | 18 | White | 68 | 1.79 | 21.1 | 9.51 | 72.37 | 1 | 9.70 | 69.22 | 5 | 11.07 | 63.36 | 25 | 12.10 | 61.52 | 31 |
| 37 | M | 23 | White | 82 | 1.89 | 22.9 | 9.86 | 47.63 | 1 | 9.47 | 48.42 | 4 | 11.93 | 45.35 | 21 | 10.18 | 39.29 | 23 |
| 38 | F | 22 | White | 69 | 1.64 | 24.5 | 11.31 | 65.20 | 1 | 12.23 | 72.45 | 5 | ex | ex | ex | ex | ex | ex |
| 39 | M | 21 | White | 71 | 1.70 | 24.4 | 10.07 | 44.78 | 1 | 9.48 | 48.56 | 1 | 10.96 | 53.65 | 21 | 10.15 | 37.65 | 21 |
| 40 | M | 22 | White | 74 | 1.75 | 21.2 | 9.97 | 56.64 | 1 | 10.29 | 60.09 | 4 | 9.53 | 50.64 | 20 | 8.67 | 52.50 | 23 |
| 41 | M | 20 | Black | 80 | 1.72 | 27.2 | 10.63 | 29.76 | 2 | 9.21 | 37.47 | 4 | 10.24 | 35.99 | 28 | 9.33 | 36.53 | 29 |
| 42 | M | 19 | White | 79 | 1.86 | 22.9 | 11.02 | 63.70 | 6 | 10.39 | 66.68 | 3 | 12.64 | 62.74 | 24 | 13.71 | 58.63 | 28 |
| 43 | M | 19 | White | 65 | 1.76 | 28.1 | 12.68 | 55.90 | 1 | 11.82 | 61.36 | 5 | ex | ex | ex | ex | ex | ex |
| 44 | F | 18 | White | 71 | 1.73 | 27.1 | 9.03 | 74.50 | 1 | 8.30 | 79.54 | 5 | ex | ex | ex | 8.34 | 64.12 | 29 |
| 45 | M | 21 | White | 90 | 1.79 | 27.3 | 9.99 | 40.59 | 1 | 9.83 | 33.21 | 4 | 9.65 | 29.78 | 28 | 11.55 | 28.59 | 33 |
| 46 | M | 21 | Black | 79 | 1.75 | 26.0 | 10.29 | 24.27 | 1 | 10.24 | 24.69 | 1 | 12.44 | 25.97 | 28 | ex | ex | ex |
| 47 | M | 21 | Other | 85 | 1.72 | 28.8 | 7.89 | 65.95 | 1 | 8.87 | 63.56 | 1 | 10.11 | 83.03 | 18 | 9.15 | 82.92 | 18 |
| 48 | M | 24 | White | 61 | 1.78 | 26.9 | 10.15 | 58.76 | 1 | 9.88 | 54.96 | 3 | 11.03 | 46.84 | 29 | 10.93 | 47.86 | 33 |
| 49 | M | 19 | White | 89 | 1.86 | 21.9 | 10.67 | 44.40 | 1 | 8.11 | 24.36 | 4 | 9.05 | 37.54 | 22 | 10.63 | 22.68 | 24 |
| 50 | M | 18 | White | 80 | 1.86 | 20.1 | 10.47 | 31.63 | 1 | 10.06 | 39.22 | 3 | 10.98 | 31.76 | 17 | 11.35 | 29.59 | 18 |
| 51 | M | 22 | White | 71 | 1.79 | 21.7 | 11.10 | 57.40 | 1 | 10.29 | 52.81 | 3 | 13.32 | 71.22 | 26 | 13.08 | 57.04 | 27 |
| 52 | M | 23 | White | 70 | 1.70 | 24.4 | 11.18 | 56.90 | 1 | 9.83 | 46.86 | 4 | 11.00 | 34.40 | 30 | 10.61 | 27.71 | 32 |
| 53 | M | 20 | White | 65 | 1.83 | 18.1 | ex | ex | ex | ex | ex | ex | 12.04 | 69.94 | 29 | ex | ex | ex |
| 54 | M | 22 | White | 82 | 1.78 | 25.9 | 13.62 | 50.01 | 1 | 12.07 | 51.42 | 4 | 12.95 | 39.23 | 27 | 11.37 | 48.26 | 31 |
| 55 | M | 21 | White | 85 | 1.79 | 26.6 | 12.77 | 52.81 | 1 | 13.04 | 48.22 | 3 | 12.49 | 41.37 | 26 | 12.14 | 37.73 | 28 |
| 56 | M | 20 | Black | 84 | 1.82 | 25.5 | 9.63 | 61.60 | 1 | 9.68 | 70.77 | 5 | 10.18 | 44.26 | 30 | 9.76 | 28.30 | 31 |
| 57 | F | 22 | White | 64 | 1.70 | 27.4 | 9.65 | 75.99 | 1 | 8.69 | 69.11 | 6 | ex | ex | ex | 9.55 | 51.65 | 32 |
| 58 | F | 22 | White | 66 | 1.75 | 24.0 | 9.92 | 48.85 | 1 | 9.80 | 58.94 | 4 | 10.32 | 57.45 | 29 | 10.71 | 44.58 | 33 |
| 59 | F | 22 | White | 73 | 1.61 | 23.5 | 8.38 | 91.58 | 2 | 8.01 | 81.73 | 2 | 9.66 | 74.55 | 29 | 9.53 | 73.28 | 33 |
| 60 | F | 19 | White | 71 | 1.75 | 22.5 | 9.10 | 88.77 | 1 | 10.01 | 88.34 | 4 | 11.71 | 81.12 | 29 | 11.93 | 77.25 | 33 |
| 61 | F | 19 | White | 84 | 1.65 | 24.6 | 9.66 | 72.55 | 1 | 8.57 | 70.69 | 1 | 12.21 | 71.13 | 32 | 11.09 | 59.52 | 32 |
| 62 | F | 22 | White | 67 | 1.59 | 19.9 | 7.91 | 74.71 | 1 | 7.59 | 63.61 | 1 | 6.56 | 78.04 | 26 | 7.32 | 73.76 | 26 |
| 63 | F | 26 | Black | 93 | 1.82 | 28.2 | 9.78 | 53.99 | 4 | 10.82 | 62.93 | 5 | 11.71 | 61.23 | 25 | 11.84 | 53.36 | 29 |
| 64 | F | 19 | White | 63 | 1.65 | 19.7 | 9.96 | 61.81 | 2 | 9.29 | 63.05 | 4 | 11.67 | 53.61 | 24 | 11.44 | 49.30 | 29 |
| 65 | F | 18 | White | 67 | 1.79 | 24.6 | 9.48 | 42.82 | 1 | 10.29 | 52.33 | 4 | 11.38 | 49.93 | 26 | ex | ex | ex |
| 66 | F | 21 | White | 65 | 1.67 | 24.2 | 7.66 | 58.52 | 1 | 8.18 | 74.94 | 4 | 7.89 | 57.10 | 22 | 8.76 | 71.43 | 26 |
| 67 | F | 22 | White | 76 | 1.70 | 24.1 | 9.70 | 73.07 | 1 | 11.34 | 75.36 | 3 | 11.73 | 64.53 | 29 | 12.37 | 54.84 | 33 |
| 68 | F | 23 | White | 62 | 1.64 | 22.8 | 8.08 | 90.48 | 1 | 8.01 | 92.05 | 1 | 8.30 | 82.17 | 25 | 7.99 | 81.22 | 25 |
| 69 | M | 21 | White | 75 | 1.74 | 25.2 | 9.78 | 47.11 | 1 | 10.71 | 54.68 | 4 | 10.76 | 41.42 | 24 | 10.20 | 36.38 | 29 |
| 70 | F | 25 | Black | 88 | 1.80 | 27.3 | 8.84 | 60.70 | 7 | 8.26 | 47.65 | 9 | 9.71 | 48.71 | 27 | 9.16 | 48.22 | 29 |
| 71 | M | 21 | White | 95 | 1.77 | 30.2 | 10.44 | 54.50 | 1 | 10.73 | 47.97 | 4 | 11.79 | 38.85 | 24 | 12.34 | 29.49 | 24 |
| 72 | M | 22 | White | 87 | 1.76 | 28.2 | 10.96 | 53.85 | 1 | 12.01 | 60.47 | 5 | 10.96 | 41.79 | 33 | 12.11 | 39.97 | 34 |
| 73 | F | 22 | White | 67 | 1.52 | 22.4 | 6.02 | 56.33 | 1 | 5.57 | 64.45 | 1 | ex | ex | ex | ex | ex | ex |
| 74 | F | 19 | White | 67 | 1.66 | 20.3 | ex | ex | ex | ex | ex | ex | 6.25 | 62.77 | 25 | 7.48 | 61.64 | 29 |
| 75 | M | 25 | White | 79 | 1.79 | 27.7 | 9.40 | 36.00 | 1 | 9.84 | 40.28 | 3 | 9.89 | 35.10 | 29 | 11.13 | 49.59 | 32 |
| 76 | F | 22 | White | 61 | 1.83 | 26.4 | 11.48 | 54.43 | 1 | 11.18 | 55.93 | 1 | 11.65 | 56.13 | 28 | 12.16 | 52.79 | 30 |
| 77 | M | 18 | White | 79 | 1.76 | 22.8 | 7.16 | 37.07 | 1 | 6.65 | 50.92 | 4 | ex | ex | ex | ex | ex | ex |
| 78 | M | 22 | White | 79 | 1.67 | 28.4 | 10.98 | 57.33 | 1 | 11.27 | 62.22 | 1 | 11.05 | 48.14 | 22 | 11.57 | 36.71 | 22 |
| 79 | M | 21 | Other | 67 | 1.76 | 27.4 | 7.76 | 59.96 | 3 | 8.22 | 57.04 | 6 | 8.93 | 40.89 | 27 | 9.60 | 32.26 | 31 |
| 80 | M | 22 | Other | 74 | 1.81 | 22.5 | 11.09 | 44.77 | 1 | 10.99 | 40.52 | 3 | ex | ex | ex | ex | ex | ex |
| 81 | M | 20 | White | 100 | 1.99 | 25.3 | 10.51 | 68.27 | 1 | 9.50 | 60.57 | 5 | 10.49 | 59.12 | 37 | 9.31 | 47.17 | 37 |
| 82 | M | 20 | White | 69 | 1.87 | 25.1 | 11.19 | 67.54 | 1 | 11.71 | 67.55 | 5 | 12.13 | 49.37 | 37 | 13.08 | 39.16 | 38 |
| 83 | M | 23 | Hispanic | 71 | 1.73 | 23.7 | 11.69 | 40.85 | 1 | 10.62 | 41.66 | 3 | ex | ex | ex | ex | ex | ex |
| 84 | M | 21 | White | 82 | 1.78 | 26.0 | 9.64 | 69.55 | 1 | 10.12 | 60.27 | 3 | 9.58 | 68.19 | 21 | 10.78 | 65.43 | 24 |
| 85 | M | 21 | Other | 88 | 1.86 | 25.5 | 8.15 | 41.32 | 1 | 10.01 | 43.23 | 3 | 9.21 | 66.75 | 22 | 10.63 | 61.21 | 24 |
| 86 | M | 22 | White | 116 | 1.87 | 33.4 | 12.04 | 78.79 | 1 | 13.02 | 71.93 | 4 | ex | ex | ex | ex | ex | ex |
| 87 | M | 23 | White | 69 | 1.77 | 27.6 | 9.98 | 51.78 | 1 | 10.34 | 54.38 | 6 | ex | ex | ex | ex | ex | 31 |
| 88 | M | 22 | Hispanic | 82 | 1.81 | 24.9 | 7.02 | 38.89 | 1 | 6.50 | 34.42 | 3 | 8.19 | 35.91 | 31 | 8.47 | 15.79 | 31 |
| 89 | M | 21 | White | 54 | 1.74 | 24.2 | 9.62 | 52.12 | 1 | 8.94 | 49.39 | 4 | 12.35 | 46.17 | 19 | 10.83 | 43.71 | 21 |
| 90 | M | 19 | White | 50 | 1.75 | 24.4 | 11.79 | 49.62 | 1 | 10.51 | 60.19 | 3 | 11.25 | 46.32 | 27 | 12.00 | 57.42 | 28 |
| 91 | M | 20 | White | 92 | 1.85 | 27.1 | 13.36 | 52.17 | 1 | 13.67 | 52.08 | 3 | 14.12 | 54.75 | 27 | 13.49 | 46.13 | 29 |
| 92 | F | 18 | White | 56 | 1.67 | 21.2 | 8.33 | 70.57 | 1 | 6.96 | 75.23 | 4 | ex | ex | ex | ex | ex | 34 |
| 93 | F | 20 | White | 61 | 1.65 | 21.2 | 8.70 | 81.65 | 1 | 9.38 | 72.52 | 4 | 9.02 | 78.68 | 26 | 9.74 | 73.75 | 26 |
| 94 | F | 21 | White | 59 | 1.78 | 22.2 | 10.30 | 89.26 | 1 | 9.28 | 83.17 | 5 | 11.39 | 83.50 | 24 | 11.07 | 69.42 | 28 |
| 95 | F | 23 | White | 67 | 1.66 | 25.8 | 10.84 | 76.40 | 7 | 10.52 | 75.90 | 7 | 12.00 | 79.22 | 30 | 12.13 | 68.23 | 30 |
| 96 | F | 21 | White | 88 | 1.69 | 22.1 | 8.86 | 70.22 | 1 | 6.90 | 70.91 | 2 | 9.13 | 62.08 | 29 | 8.68 | 62.26 | 33 |
| 97 | F | 18 | White | 70 | 1.66 | 25.0 | 9.30 | 88.81 | 1 | 8.70 | 86.68 | 3 | 16.07 | 88.61 | 26 | ex | ex | ex |
| 98 | M | 21 | White | 58 | 1.89 | 23.7 | 11.08 | 39.14 | 1 | ex | ex | ex | ex | ex | ex | ex | ex | ex |
| 99 | M | 21 | White | 69 | 1.85 | 23.5 | 13.38 | 30.75 | 1 | 13.29 | 32.00 | 6 | 11.93 | 27.82 | 20 | 11.97 | 25.24 | 22 |
| 100 | F | 19 | White | 66 | 1.61 | 25.4 | 8.51 | 54.79 | 1 | 8.85 | 41.66 | 1 | 10.66 | 70.83 | 24 | 11.72 | 53.73 | 24 |
| 101 | F | 24 | White | 72 | 1.73 | 24.0 | ex | ex | ex | 12.54 | 79.20 | 6 | 11.29 | 53.02 | 29 | 10.15 | 61.42 | 35 |
| 102 | M | 20 | White | 102 | 1.82 | 30.9 | 10.69 | 77.85 | 1 | 10.73 | 74.16 | 1 | 10.71 | 88.79 | 26 | 8.69 | 81.02 | 26 |
| 103 | F | 23 | White | 86 | 1.64 | 24.0 | 7.47 | 92.36 | 1 | 8.85 | 89.39 | 1 | ex | ex | ex | ex | ex | ex |
| 104 | M | 23 | White | 87 | 1.82 | 26.1 | ex | ex | ex | ex | ex | ex | 10.85 | 50.18 | 23 | 11.60 | 54.79 | 26 |
| 105 | M | 22 | White | 73 | 1.79 | 22.9 | 9.96 | 63.73 | 1 | 9.99 | 56.18 | 3 | ex | ex | ex | ex | ex | 27 |
| 106 | F | 24 | White | 65 | 1.72 | 22.0 | 10.44 | 64.07 | 1 | 10.59 | 67.87 | 5 | 12.75 | 62.45 | 24 | 11.00 | 61.03 | 29 |
| 107 | M | 20 | Black | 96 | 1.76 | 31.1 | 10.58 | 23.84 | 1 | 9.36 | 30.74 | 1 | 10.86 | 25.85 | 23 | 11.92 | 38.00 | 23 |
| 108 | M | 21 | White | 84 | 1.85 | 24.6 | 9.92 | 59.90 | 1 | 10.98 | 62.37 | 4 | 9.89 | 66.62 | 27 | 11.08 | 62.32 | 28 |
| 109 | M | 22 | White | 65 | 1.78 | 25.8 | 9.32 | 41.32 | 1 | 9.94 | 37.75 | 4 | 9.56 | 24.38 | 29 | 10.55 | 33.94 | 33 |
ex = Excluded data
US image acquisition
The 109 athletes underwent a US procedure to obtain LM images at the L5 level in both the prone and standing positions. The LOGIQ e ultrasound machine (GE Healthcare, Milwaukee, WI) was used with a curvilinear probe with its imaging parameters maintained at the following values for all image acquisitions: frequency: 5 MHz, gain: 60, depth: 8.0 cm [39]. Only the LM muscle was assessed, as it is the most commonly examined muscle amongst the paraspinal muscle group using US and is the most sensitive to spinal pathology. All data collection was performed by one of the investigators (M.F.) who applied a consistent and repeated technique throughout all image acquisitions: pressure was maintained on the adjacent hand and forearm handling the probe so as to prevent tissue deformation on the region of interest through transducer pressure. The acquisition of images in the prone position consisted in having the subjects lie in the prone position on a therapy table with a pillow underneath their abdomen to decrease lumbar lordosis [8]. To assess LM CSA, transverse US images were obtained bilaterally. For subjects with larger muscles, the right and left sides were imaged unilaterally. Similarly, LM CSA measurements were obtained in the standing position, where subjects stood in their habitual standing posture [39]. The images were stored as separate datasets for each subject in *.tif format.
US image segmentation
The ground truth segmentations of LM CSA and LM EI measurements in prone and standing positions were performed on the acquired data using Fiji, a distribution of the ImageJ image processing software [40].
The ground truth segmentations for all measurements were manually obtained by one of the investigators (C.B.) who in preparation for this study, received training from another investigator (M.F.) with over 10 years of experience in spine imaging analysis. The inter-rater reliability between both investigators was examined on a set of 18 images and interclass correlation coefficient (ICC 2,1) varied between 0.93-0.99. Images of subjects where the characteristic structures and landmarks of the LM could not be clearly distinguished were excluded from the database. All ground truth segmentations for each subject are available as binary masks and stored as separate *.tif files.
Utility and discussion
Database availability
The database is available at http://data.sonography.ai. The B-mode images and binary segmentation masks for each subject are deposited as *.tif files.
Data organisation & file naming conventions
The database separates the B-mode images of each subject into a folder named “B-mode” and the masks into a folder named “Masks”. The datasets of subjects and corresponding binary segmentation masks are labelled with the same subject ID (1 to 109). The best available images (e.g. frames) for each subject were chosen for the segmentations. Since images were acquired bilaterally in some cases and unilaterally in subjects with larger muscles, different file naming conventions were used for the B-mode images as well as their corresponding masks. Table 1 can be used to verify whether a frame corresponds to either the right or left side, as well as whether the frame is in the prone or standing position.
Unilateral file naming conventions
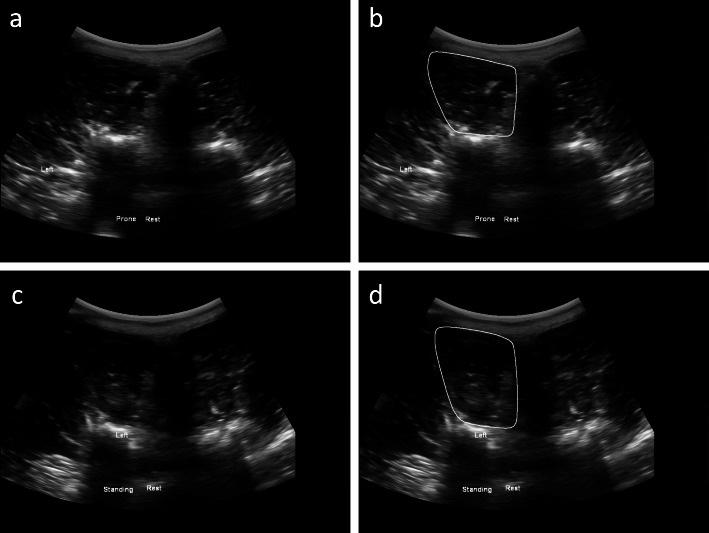
For the subjects where the images were acquired unilaterally, the B-mode images and masks have a one-to-one correspondence. The file names for the B-mode images and masks have the following generic format: X_Y_Bmode.tif and X_Y_Mask.tif, where X is the subject ID, and Y is the frame number. As an example, 50_3_Bmode.tif would have a corresponding mask 50_3_Mask.tif. This can be seen in Fig. 1a and b.
Fig. 1.
(a) B-mode image of subject 50 (acquired unilaterally) with corresponding segmentation of the left MF in the prone position shown in (b). (c) B-mode image of subject 50 (acquired unilaterally) with corresponding segmentation of the left MF in the standing position shown in (d)
Bilateral file naming conventions
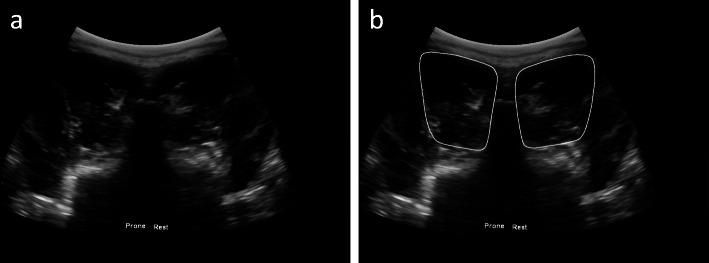
For the subjects where images were acquired bilaterally, the file names for the B-mode images and masks have the following generic format: X_Y_Bmode.tif and X_Y_MaskZ.tif, where X is the subject ID, Y is the frame number, and Z is a value of 1 or 2 used as an identifier to distinguish between the right and left side, respectively. As an example, 46_1_Bmode.tif would have corresponding masks 46_1_Mask1.tif and 46_1_Mask2.tif. This can be seen in Fig. 2a and b.
Fig. 2.
(a) B-mode image of subject 46 (acquired bilaterally) with corresponding segmentations of the left and right MF in the prone position shown in (b)
Discussion
Due to portability, cost-effectiveness, and efficiency, clinicians and researchers widely use US as an imaging modality in their screening and diagnostic procedures over other imaging modalities such as MRI, CT and X-ray. However, US presents its own set of disadvantages relating to the task of manual segmentation. Due to speckle noise in US images, manual segmentation is highly rater-dependent and thus, is susceptible to errors which affect LM analysis and results. As such, the development of powerful segmentation algorithms can help mitigate the aforementioned issues. Deep learning techniques can be employed to extract features from the data and can then be used to perform automatic US image segmentation [41]. Although potential applications of deep learning algorithms have been demonstrated for MRI and microscopy modalities, very few have focused on algorithms applied to US [37]. Furthermore, the performance of deep learning algorithms is highly dependent on a high volume of quality data. The availability of public repositories on clinical data pertaining to LM muscle images are scarce, and thus greatly limit the development and testing of the segmentation algorithms. As such, our aim with this study was to provide the first publicly available US database of the LM.
This database is comprised of 109 subjects with the ground truth of the left and right LM at the L5 level in both prone and standing positions. The ground truth data can enable the development of deep learning algorithms used for automatic segmentation tasks related to the LM. Given the volume of the annotated data, the developed algorithm can have a better generalization capability through proper parameter tuning and data augmentation [41]. Moreover, deep learning algorithms can exploit the morphological features that trained experts use to perform their segmentations [41, 42]. Furthermore, the algorithms can produce comparable results to those of the examiner [36, 43]. As such, examiner subjectivity during assessment of muscle morphology can be reduced. In addition, it would greatly benefit clinicians and researchers whilst enabling them to perform assessments in a practical and time-efficient manner.
This database contributes and dedicates itself to advancing the development of automatic segmentation algorithms related to the assessment of LM muscle morphology. However, this database only includes young athletic adults aged between 18 and 26 years old. Within the dataset, there are natural variances in age, BMI, and other underlying conditions which may differ from one participant to the next. As such, algorithms which are developed using our dataset should be mindful of these limitations and foresee difficulties in accurate segmentation when subjected to samples from other populations, a problem commonly referred to as domain shift. Thus, future efforts need to be made to extend this database to include sedentary and older adults, which are more representative of the general population suffering from LBP. When viewing US image of younger muscle (e.g. higher fluid content), contrasting echogenicities of hypoechoic (toward black) muscle and hyperechoic (toward white) fascia allow for easier tissue differentiation and identification of key landmarks [44]. With ageing, there is a natural increase in fibrous tissue and thus the distinction between muscle and fascia is more difficult [25, 45]. As such, this database should be treated as a platform of an ongoing process towards the automatization and standardization of LM muscle measurements from US images.
Conclusion
Herein, we presented the LUMINOUS database which contains manual segmentations of LM images at the L5 level obtained via US as well as their corresponding binary masks. The database is comprised of 109 datasets, which will enable the development of automated segmentation algorithms of the LM. This database will provide a means to support the standardization of US measurements, facilitate comparison between studies and accelerate the clinical implementation of quantitative muscle assessment in clinical and research settings.
Acknowledgements
The authors would like to thank Ms. Nadia El Hallaoui for her valuable discussions.
Abbreviations
- US
Ultrasound
- LM
Lumbar multifidus
- CSA
Cross-sectional area
- LBP
Lower back pain
- MRI
Magnetic resonance imaging
- CT
Computed tomography
- EI
Echo intensity
- BMI
Body mass index
Authors’ contributions
Conception and design: CB, MF, HR, MB. Analysis and interpretation of data: CB, MF. Collection and assembly of data: CB, BB. Drafting of the manuscript: CB, BB, MF. Critical revision of the manuscript: MB, HR, HB. Funding: MF, HR, HB. Final approval of the manuscript: CB, BB, HR, MB, HB, MF. All authors read and approved the final version of the manuscript.
Funding
The present work was supported by the PERFORM Centre (Concordia University), the R. Howard Webster Foundation, and the Natural Sciences and Engineering Research Council of Canada (NSERC) RGPIN-2020-04612.
Availability of data and materials
The database presented in this study is freely available online at http://data.sonography.ai.
Ethics approval and consent to participate
This study was approved by the Concordia University Human Research Ethics Committee (certification number: 30011880). Written informed consent to participate in the study was obtained from each participant.
Consent for publication
Written informed consent to publish data in the study was obtained from each participant.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Clyde J. Belasso, Email: clyde.belasso@gmail.com
Bahareh Behboodi, Email: behboodibahareh@gmail.com.
Habib Benali, Email: habib.benali@concordia.ca.
Mathieu Boily, Email: matt_boily@hotmail.com.
Hassan Rivaz, Email: hrivaz@ece.concordia.ca.
Maryse Fortin, Email: maryse.fortin@concordia.ca.
References
- 1.Preston DC, Shapiro BE. 29 - Radiculopathy. In: Preston DC, Shapiro BE, editors. Electromyography and Neuromuscular Disorders. London: W B Saunders; 2013. [Google Scholar]
- 2.Stokes M, Hides J, Elliott J, Kiesel K, Hodges P. Rehabilitative ultrasound imaging of the posterior paraspinal muscles. J Orthop Sports Phys Ther. 2007;37(10):581–95. doi: 10.2519/jospt.2007.2599. [DOI] [PubMed] [Google Scholar]
- 3.Wilke H-J, Wolf S, Claes LE, Arand M, Wiesend A. Stability increase of the lumbar spine with different muscle groups. a biomechanical in vitro study. Spine. 1995;20(2):192–7. doi: 10.1097/00007632-199501150-00011. [DOI] [PubMed] [Google Scholar]
- 4.Freeman MD, Woodham MA, Woodham AW. The role of the lumbar multifidus in chronic low back pain: A review. PM&R. 2010;2(2):142–6. doi: 10.1016/j.pmrj.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Panjabi MM. The stabilizing system of the spine. Part II. Neutral zone and instability hypothesis. J Spinal Disord. 1992;5(2):390–7. doi: 10.1097/00002517-199212000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Macintosh JE, Valencia F, Bogduk N, Munro RR. The morphology of the human lumbar multifidus. Clin Biomech. 1986;1(4):196–204. doi: 10.1016/0268-0033(86)90146-4. [DOI] [PubMed] [Google Scholar]
- 7.Ward SR, Kim CW, Eng CM, Gottschalk IV LJ, Tomiya A, Garfin SR, Lieber RL. Architectural analysis and intraoperative measurements demonstrate the unique design of the multifidus muscle for lumbar spine stability. J Bone Joint Surg Am Vol. 2009;91(1):176–85. doi: 10.2106/JBJS.G.01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skeie EJ, Borge JA, Leboeuf-Yde C, Bolton J, Wedderkopp N. Reliability of diagnostic ultrasound in measuring the multifidus muscle. Chiropr Man Ther. 2015;23(1):15. doi: 10.1186/s12998-015-0059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suni J, Rinne M, Natri A, Statistisian MP, Parkkari J, Alaranta H. Control of the lumbar neutral zone decreases low back pain and improves self-evaluated work ability: a 12-month randomized controlled study. Spine. 2006;31(18):611–20. doi: 10.1097/01.brs.0000231701.76452.05. [DOI] [PubMed] [Google Scholar]
- 10.Aure OF, Nilsen JH, Vasseljen O. Manual therapy and exercise therapy in patients with chronic low back pain: a randomized, controlled trial with 1-year follow-up. Spine. 2003;28(6):525–31. doi: 10.1097/01.BRS.0000049921.04200.A6. [DOI] [PubMed] [Google Scholar]
- 11.Buchbinder R, van Tulder M, Öberg B, Costa LM, Woolf A, Schoene M, Croft P, Hartvigsen J, Cherkin D, Foster NE, Maher CG. Low back pain: a call for action. The Lancet. 2018;391(10137):2384–8. doi: 10.1016/S0140-6736(18)30488-4. [DOI] [PubMed] [Google Scholar]
- 12.Foster NE, Anema JR, Cherkin D, Chou R, Cohen SP, Gross DP, Ferreira PH, Fritz JM, Koes BW, Peul W, Turner JA. Prevention and treatment of low back pain: evidence, challenges, and promising directions. The Lancet. 2018;391(10137):2368–83. doi: 10.1016/S0140-6736(18)30489-6. [DOI] [PubMed] [Google Scholar]
- 13.Ranger TA, Cicuttini FM, Jensen TS, Peiris WL, Hussain SM, Fairley J, Urquhart DM. Are the size and composition of the paraspinal muscles associated with low back pain? A systematic review. Spine J. 2017;17(11):1729–48. doi: 10.1016/j.spinee.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Shahidi B, Hubbard JC, Gibbons MC, Ruoss S, Zlomislic V, Allen RT, Garfin SR, Ward SR. Lumbar multifidus muscle degenerates in individuals with chronic degenerative lumbar spine pathology. J Orthop Res. 2017;35(12):2700–6. doi: 10.1002/jor.23597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodges PW, Danneels L. Changes in structure and function of the back muscles in low back pain: different time points, observations, and mechanisms. J Orthop Sports Phys Ther. 2019;49(6):464–76. doi: 10.2519/jospt.2019.8827. [DOI] [PubMed] [Google Scholar]
- 16.Prins MR, Griffioen M, Veeger TT, Kiers H, Meijer OG, van der Wurff P, Bruijn SM, van Dieën JH. Evidence of splinting in low back pain? A systematic review of perturbation studies. Eur Spine J. 2018;27:40–59. doi: 10.1007/s00586-017-5287-0. [DOI] [PubMed] [Google Scholar]
- 17.Fortin M, Lazáry À, Varga PP, Battié MC. Association between paraspinal muscle morphology, clinical symptoms and functional status in patients with lumbar spinal stenosis. Eur Spine J. 2017;26(10):2543–51. doi: 10.1007/s00586-017-5228-y. [DOI] [PubMed] [Google Scholar]
- 18.Fortin M, Gibbons L, Videman T, Battié M. Do variations in paraspinal muscle morphology and composition predict low back pain in men? Scand J Med Sci Sports. 2015;25(6):880–7. doi: 10.1111/sms.12301. [DOI] [PubMed] [Google Scholar]
- 19.Hildebrandt M, Fankhauser G, Meichtry A, Luomajoki H. Correlation between lumbar dysfunction and fat infiltration in lumbar multifidus muscles in patients with low back pain. BMC Musculoskelet Disord. 2017;18:12. doi: 10.1186/s12891-016-1376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teichtahl AJ, Urquhart DM, Wang Y, Wluka AE, Wijethilake P, O’Sullivan R, Cicuttini FM. Fat infiltration of paraspinal muscles is associated with low back pain, disability, and structural abnormalities in community-based adults. Spine J. 2015;15(7):1593–601. doi: 10.1016/j.spinee.2015.03.039. [DOI] [PubMed] [Google Scholar]
- 21.Khan AB, Weiss EH, Khan AW, Omeis I, Verla T. Back muscle morphometry: effects on outcomes of spine surgery. World Neurosurg. 2017;103:174–9. doi: 10.1016/j.wneu.2017.03.097. [DOI] [PubMed] [Google Scholar]
- 22.Storheim K, Berg L, Hellum C, Gjertsen Ø, Neckelmann G, Espeland A, Keller A, Group NSS, et al. Fat in the lumbar multifidus muscles-predictive value and change following disc prosthesis surgery and multidisciplinary rehabilitation in patients with chronic low back pain and degenerative disc: 2-year follow-up of a randomized trial. BMC Musculoskelet Disord. 2017;18:145. doi: 10.1186/s12891-017-1505-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frantz Pressler J, Givens Heiss D, Buford JA, Chidley JV. Between-day repeatability and symmetry of multifidus cross-sectional area measured using ultrasound imaging. J Orthop Sports Phys Ther. 2006;36(1):10–8. doi: 10.2519/jospt.2006.36.1.10. [DOI] [PubMed] [Google Scholar]
- 24.Mehyar F, Santos M, Wilson SE, Staggs VS, Sharma NK. Immediate effect of lumbar mobilization on activity of erector spinae and lumbar multifidus muscles. J Chiropr Med. 2017;16(4):271–8. doi: 10.1016/j.jcm.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sions JM, Velasco TO, Teyhen DS, Hicks GE. Ultrasound imaging: Intraexaminer and interexaminer reliability for multifidus muscle thickness assessment in adults aged 60 to 85 years versus younger adults. J Orthop Sports Phys Ther. 2014;44(6):425–34. doi: 10.2519/jospt.2014.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuellar WA, Blizzard L, Callisaya ML, Hides JA, Jones G, Ding C, Winzenberg TM. Test-retest reliability of measurements of abdominal and multifidus muscles using ultrasound imaging in adults aged 50–79 years. Musculoskelet Sci Pract. 2017;28:79–84. doi: 10.1016/j.msksp.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Koppenhaver SL, Hebert JJ, Fritz JM, Parent EC, Teyhen DS, Magel JS. Reliability of rehabilitative ultrasound imaging of the transversus abdominis and lumbar multifidus muscles. Arch Phys Med Rehabil. 2009;90(1):87–94. doi: 10.1016/j.apmr.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 28.Djordjevic O, Djordjevic A, Konstantinovic L. Interrater and intrarater reliability of transverse abdominal and lumbar multifidus muscle thickness in subjects with and without low back pain. J Orthop Sports Phys Ther. 2014;44(12):979–88. doi: 10.2519/jospt.2014.5141. [DOI] [PubMed] [Google Scholar]
- 29.Caresio C, Molinari F, Emanuel G, Minetto MA. Muscle echo intensity: reliability and conditioning factors. Clin Physiol Funct Imaging. 2015;35(5):393–403. doi: 10.1111/cpf.12175. [DOI] [PubMed] [Google Scholar]
- 30.Pillen S, Tak RO, Zwarts MJ, Lammens MM, Verrijp KN, Arts IM, van der Laak JA, Hoogerbrugge PM, van Engelen BG, Verrips A. Skeletal muscle ultrasound: correlation between fibrous tissue and echo intensity. Ultrasound Med Biol. 2009;35(3):443–6. doi: 10.1016/j.ultrasmedbio.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Arts IM, Pillen S, Schelhaas HJ, Overeem S, Zwarts MJ. Normal values for quantitative muscle ultrasonography in adults. Muscle Nerve Off J Am Assoc Electrodiagnostic Med. 2010;41(1):32–41. doi: 10.1002/mus.21458. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe Y, Yamada Y, Fukumoto Y, Ishihara T, Yokoyama K, Yoshida T, Miyake M, Yamagata E, Kimura M. Echo intensity obtained from ultrasonography images reflecting muscle strength in elderly men. Clin Interventions Aging. 2013;8:993. doi: 10.2147/CIA.S47263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranson CA, Burnett AF, Kerslake R, Batt ME, O’Sullivan PB. An investigation into the use of MR imaging to determine the functional cross sectional area of lumbar paraspinal muscles. Eur Spine J. 2006;15:764–73. doi: 10.1007/s00586-005-0909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Luo H, Liu Y. Paraspinal muscle segmentation based on deep neural network. Sensors. 2019;19(12):2650. doi: 10.3390/s19122650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burian E, Rohrmeier A, Schlaeger S, Dieckmeyer M, Diefenbach MN, Syväri J, Klupp E, Weidlich D, Zimmer C, Rummeny EJ, et al. Lumbar muscle and vertebral bodies segmentation of chemical shift encoding-based water-fat MRI: the reference database myosegmentum spine. BMC Musculoskelet Disord. 2019;20:152. doi: 10.1186/s12891-019-2528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber KA, Smith AC, Wasielewski M, Eghtesad K, Upadhyayula PA, Wintermark M, Hastie TJ, Parrish TB, Mackey S, Elliott JM. Deep learning convolutional neural networks for the automatic quantification of muscle fat infiltration following whiplash injury. Sci Rep. 2019;9(1):1–8. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, Wang Y, Yang X, Lei B, Liu L, Li SX, Ni D, Wang T. Deep learning in medical ultrasound analysis: a review. Engineering. 2019;5(2):261–75. doi: 10.1016/j.eng.2018.11.020. [DOI] [Google Scholar]
- 38.Cunningham RJ, Harding PJ, Loram ID. Real-time ultrasound segmentation, analysis and visualisation of deep cervical muscle structure. IEEE Trans Med Imaging. 2016;36(2):653–65. doi: 10.1109/TMI.2016.2623819. [DOI] [PubMed] [Google Scholar]
- 39.Nandlall N, Rivaz H, Rizk A, Frenette S, Boily M, Fortin M. The effect of low back pain and lower limb injury on lumbar multifidus muscle morphology and function in university soccer players. BMC Musculoskelet Disord. 2020;21:96. doi: 10.1186/s12891-020-3119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haque IRI, Neubert J. Deep learning approaches to biomedical image segmentation. Inform Med Unlocked. 2020;18:100297. doi: 10.1016/j.imu.2020.100297. [DOI] [Google Scholar]
- 42.Park J, Kim DI, Choi B, Kang W, Kwon HW. Classification and morphological analysis of vector mosquitoes using deep convolutional neural networks. Sci Rep. 2020;10(1):1012. doi: 10.1038/s41598-020-57875-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salvi M, Caresio C, Meiburger KM, De Santi B, Molinari F, Minetto MA. Transverse muscle ultrasound analysis (TRAMA): Robust and accurate segmentation of muscle cross-sectional area. Ultrasound Med Biol. 2019;45(3):672–83. doi: 10.1016/j.ultrasmedbio.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Whittaker JL, Teyhen DS, Elliott JM, Cook K, Langevin HM, Dahl HH, Stokes M. Rehabilitative ultrasound imaging: understanding the technology and its applications. J Orthop Sports Phys Ther. 2007;37(8):434–49. doi: 10.2519/jospt.2007.2350. [DOI] [PubMed] [Google Scholar]
- 45.Stokes M, Rankin G, Newham D. Ultrasound imaging of lumbar multifidus muscle: normal reference ranges for measurements and practical guidance on the technique. Man Ther. 2005;10(2):116–26. doi: 10.1016/j.math.2004.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The database presented in this study is freely available online at http://data.sonography.ai.