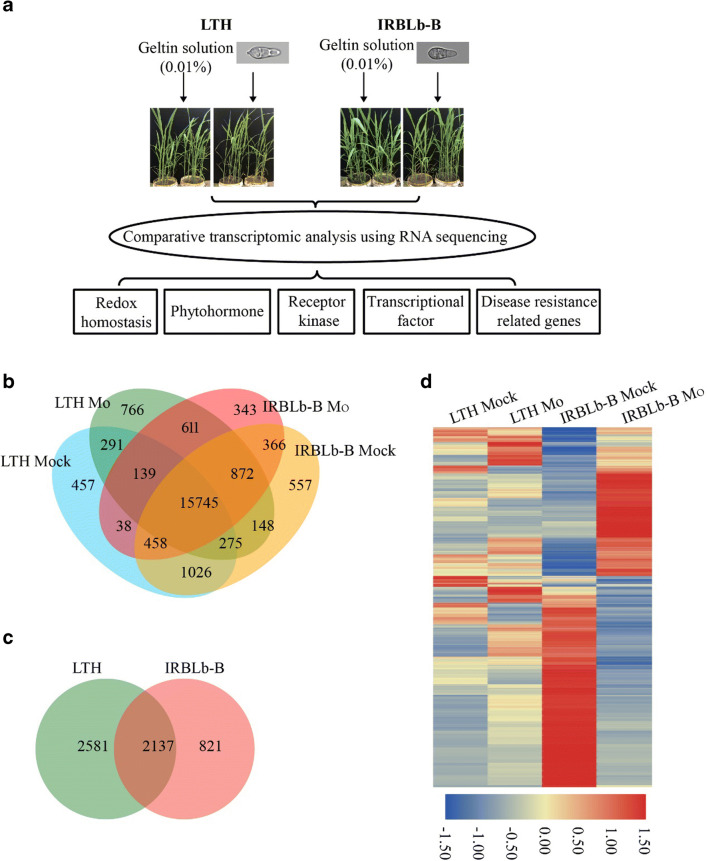
Fig. 2.
The RNA-seq workflow and global profiling. a The RNA-seq workflow of IRBLb-B and LTH with M. oryzae strain P131 infection. Redox homeostasis, phytohormone, receptor kinase, transcriptional factor, and disease resistance related genes were analyzed to reveal the Pib-mediated blast resistance. b The RNA-seq based identification of differential transcription in seedlings of IRBLb-B and LTH sampled 36 hpi with M. oryzae strain P131. Venn diagram illustrating the number of genes detected present in both genotypes and/or treatments, and those present in specific genotype and/or treatment combinations. c The venn diagram of SDEGs (FDR adjusted p ≤ 0.05 and absolute log2 fold ≥ 2) across LTH and IRBLb-B. d The heatmap of SDEGs across LTH and IRBLb-B