Fig. 2.
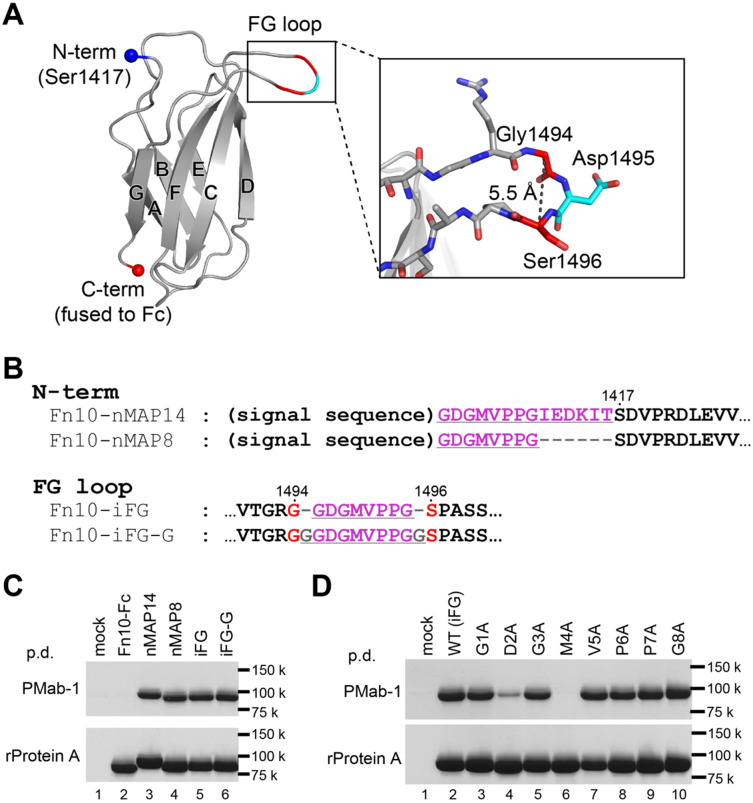
The MAP tag can be inserted into the long protruded loop of Fn10. (A) Fn10 structure extracted from the crystal structure of human fibronectin fragment (PDB ID: 1fnf) is shown as a cartoon model. The N- and C-termini of Fn10 are shown as spheres. The seven strands forming the β-sandwich structure of Fn10 are labelled with A–G. The expanded view of the FG loop shown as stick models is provided in the inset. (B) The amino-acid sequences for N-terminally MAP-tagged or MAP-inserted Fn10-Fc mutants near the MAP tag-fused portions. The MAP tag-derived sequences and linker residues are underlined. Residue numbers of original Fn10 are provided above the sequences. (C and D) Pull-down assay of the MAP-tagged Fn10-Fc by PMab-1-immobilized Sepharose (upper panel) and rProtein A Sepharose (lower panel). The Fn10-Fc samples were expressed in Expi293F cells and precipitated with each resin, followed by 12.5% SDS–PAGE under non-reducing conditions and stained with Coomassie Brilliant Blue. All mutants were expressed well in Expi293F cells as confirmed by the rProtein A Sepharose pull-down assay (C, D, lower panels). Note that the nMAP8, iFG and iFG-G samples were captured by PMab-1 with equivalent efficiency with the nMAP14 sample (C, top panel) and that the D2A and M4A mutants showed severe decrease in binding affinity for PMab-1 (D, top panel).