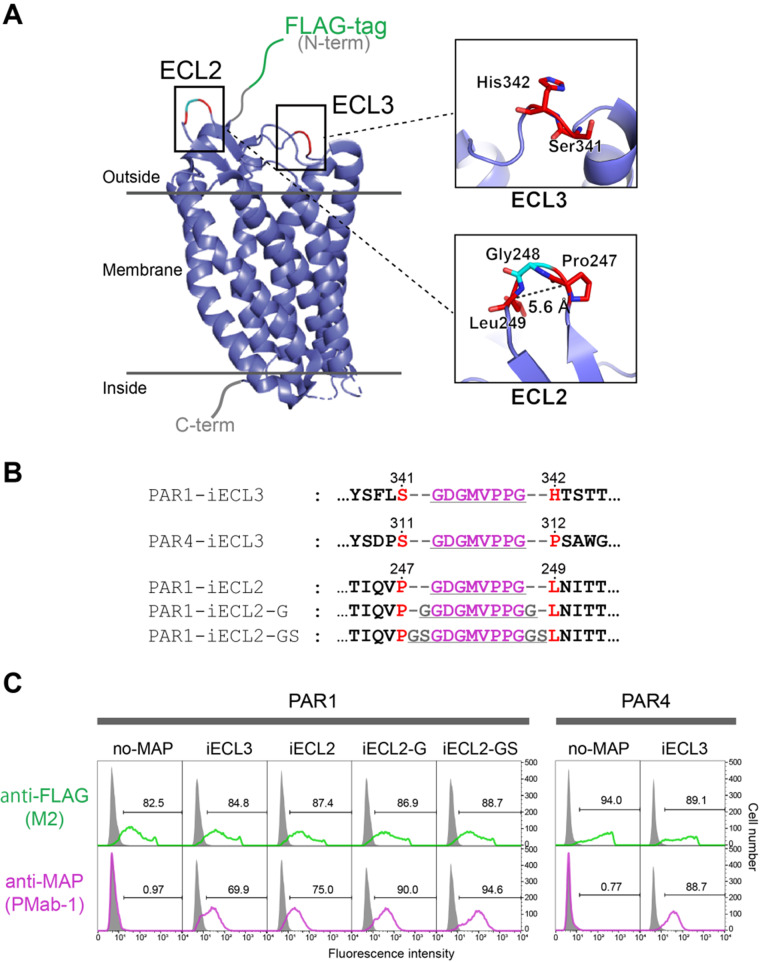
Fig. 3.
The MAP-inserted GPCRs expressing on human cells were efficiently stained with PMab-1 in flow cytometry analysis. (A) Overall structure of PAR1. The crystal structure of PAR1 was determined as a T4 lysozyme-fused mutant (PDB ID: 3vw7), and the figure was drawn as a cartoon model after eliminating the T4 lysozyme portion from the model. The expanded view at the MAP tag insertion regions is shown in the right. (B) The amino-acid sequences for MAP-inserted PAR1 and PAR4 mutants near the insertion portions. The MAP8 tag sequence and linker residues are underlined. Residue numbers of the original receptors are provided above the sequences. (C) Flow cytometry analysis of MAP-inserted PAR1 and PAR4 mutants. PAR1 and PAR4 mutants were transiently expressed on Expi293F cells and incubated with PMab-1 or anti-FLAG antibody M2, followed by staining with corresponding Alexa-Fluor-488-labelled secondary antibodies. Signals from cells transfected with each sample and not treated with any antibodies (grey area) were used to define PMab-1- or FLAG-positive cell populations (regions including <1% of non-treated cells were indicated by the brackets and defined as the positive region for each antibody). Staining efficiencies are expressed as the % positive cells and provided in the each panel.