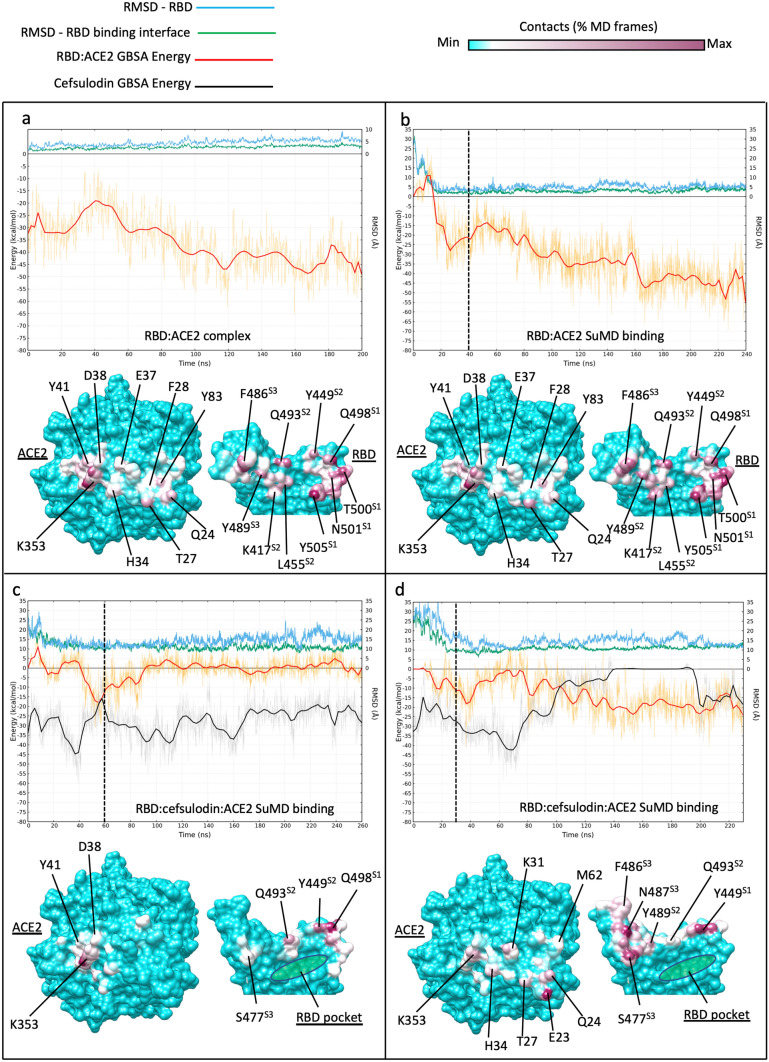
Fig. 3.
Influence of RBD-bound cefsulodin on the intermolecular recognition between RBD and ACE2 during binding. Top panels plots show the RMSD with respect to PDB 6M17 of the whole RBD (cyan line) and the RBD residues located at the binding interface with ACE2 (green line), respectively; the red and black lines indicate the MMGBSA binding energy of RBD:ACE2 complex and cefsulodin, respectively. Bottom panels: RBD:ACE2 intermolecular contacts potted on the surface of the proteins; the RBD pocket, which was occupied by cefsulodin at the beginning of the simulations, is indicated in green in (c) and (d). a classic MD simulation of the RBD:ACE2 complex from PDB 6M17; b RBD:ACE SuMD binding; the dimer reached the 6M17 bound conformation (RMSD ≈ 2 Å) in less than 20 ns. c)) RBD:cefsulodin:ACE ternary complex SuMD binding replica 1; despite reaching the ACE2 surface, RBD was not stabilized due to the co-presence of cefsulodin on protein interfaces; d RBD:cefsulodin:ACE2 ternary complex SuMD binding replica 2; cefsulodin was displaced by ACE2 after 140 ns of simulation, However, the RBD did not reach the experimental conformation within 230 ns (RMSD ≈ 10–15 Å).). Vertical dashed lines indicate the start of 200 ns of classic MD after SuMD; the three RBD sites responsible for binding ACE2 are indicated with superscripts on the RBD residue names (S1, S2, and S3)