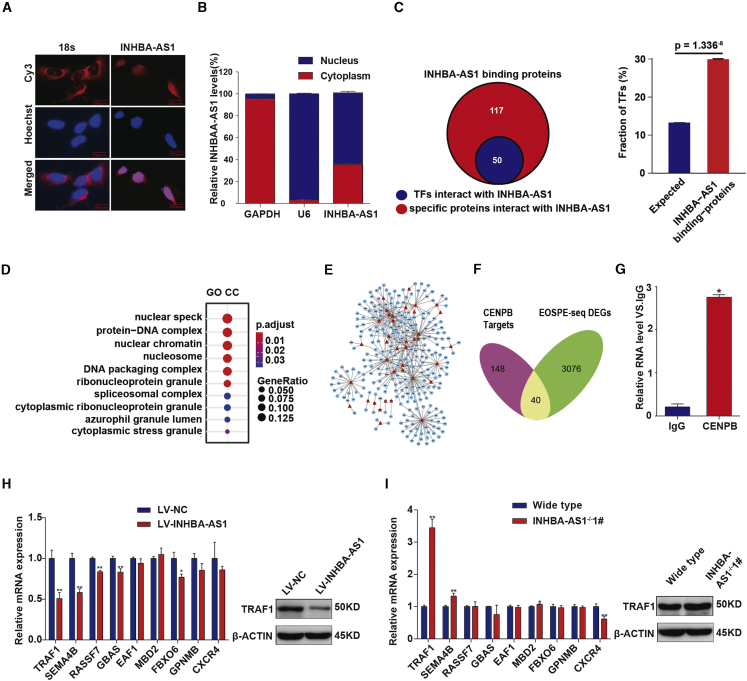
Figure 4.
INHBA-AS1 Interacts with CENPB and Inhibits TRAF1 Expression
(A) The subcellular localization of INHBA-AS1 in HTR-8/SVneo detected by FISH assays. Red, INHBA-AS1; blue, nucleus. Scale bar, 20 μm. (B) The subcellular localization of INHBA-AS1 in the HTR-8/SVneo cell detected by cell fractionation assays. U6, nucleus marker; GAPDH, cytoplasm marker. (C) INHBA-AS1-binding proteins detected by pulldown and mass spectrometry. (D) Enrichment analysis of GO term cellular component (GO-CC) on INHBA-AS1-binding proteins. (E) TF-target network (TFTNet) for EOSPE DEGs. Red triangles, TFs; blue nodes, targets of the TFs. (F) Overlaps between CENPB target genes (188) and DEGs (3,116) in EOSPE detected in transcriptome sequencing. (G) Binding of INHBA-AS1 with CENPB detected by RIP assays and qRT-PCR. (H) Transcription level of CENPB target genes in cells with INHBA-AS1 overexpression. Left panel shows qPCR results; blue bars, cells transfected with empty vector; red bars, cells transfected with INHBA-AS1; right panel shows western blotting results for TRAF1. (I) Transcription level detected of CENPB target genes in cells with INHBA-AS1 knockout. Left panel shows qPCR results; blue bars, cells without knockout; red bars, cells with INHBA-AS1 knockout; right panel shows western blotting result of TRAF1. The values are shown as the mean ± SD of three independent experiments; ∗∗p < 0.01, ∗p < 0.05. See also Figure S2 and Tables S2, S3, and S4.