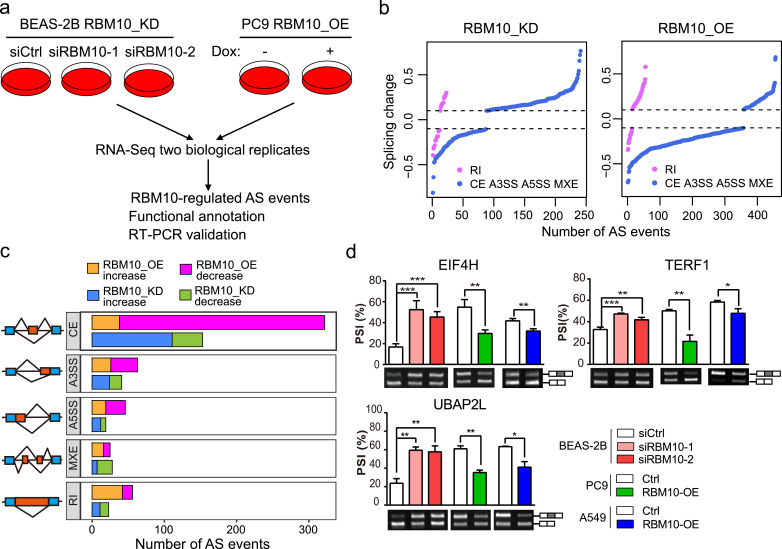
Fig. 4.
RBM10 regulates alternative splicing (AS) of cancer-associated genes. (a) Experimental design for identifying AS changes induced by RBM10 knockdown (RBM10_KD) in lung epithelial BEAS-2B cells and overexpression (RBM10_OE) in LUAD PC9 cells. (b) Splicing changes following RBM10 KD in BEAS-2B cells or RBM10 OE in PC9 cells (P < 0.05, |splicing change| > 0.1). AS types were indicated in Fig. 2b. (c) The numbers of altered splicing events in each AS category. (d) RT-PCR validation of RBM10-regulated AS events under the indicated conditions. Agarose gel images (lower panel) and quantification of exon inclusion levels (upper panel) are shown for 3 representative genes. PSI: percent-spliced-in. Error bars represent +SEM, n = 3–4 biological replicates; * P < 0.05, ** P < 0.01, *** P < 0.001; One-way ANOVA followed by Dunnett's tests were used for comparisons with control (siCtrl) in BEAS-2B cells; Student's t-tests were used for comparisons with control (Ctrl) in PC9 and A549 cells.