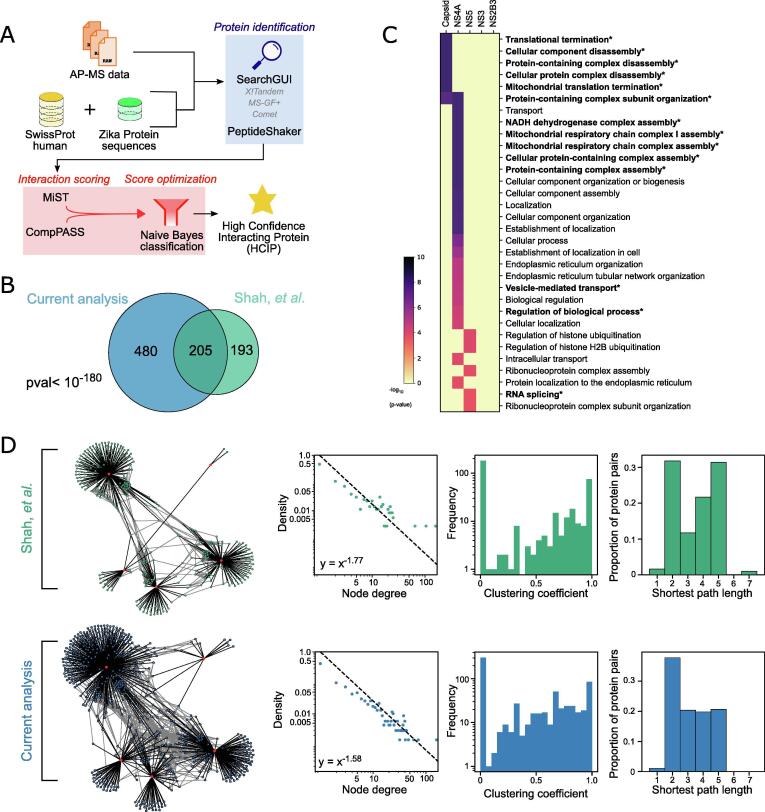
Fig. 1.
Evaluating reproducibility across AP-MS data analysis A. Graphical representation of the pipeline used for affinity-purification mass spectrometry (AP-MS) data analysis. AP-MS data is interrogated using a combined database of human protein sequences (yellow) and Zika viral protein sequences (green). The pipeline contains a step of protein identification (blue) using SearchGUI and PeptideShaker, followed by interaction scoring (red) using MiST, CompPASS and a Naïve Bayes classification algorithm, to produce a list of highly confident interacting proteins (HCIPs). B. Overlap of proteins identified in this study (current analysis) with that of the original study from Shah and colleagues [8]. Significance of the overlap was evaluated using a Fisher’s exact test. C. Gene ontology (GO) enrichment of interacting proteins (preys) for each viral proteins (baits – top). The heatmap displays all significant enrichments (FDR < 1%, Benjamini-Hochberg correction). The GO terms highlighted in bold and a star correspond to GO terms also identified in the original study. D. Network characteristics between the current analysis and that of the original study. Networks are represented with bait proteins in red (Zika proteins) and interacting proteins in green for the original study and in blue for the current analysis. The plots from left to right correspond to the node degree distribution, the distribution of local clustering coefficients and the distribution of shortest pair length. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)