Figure 1.
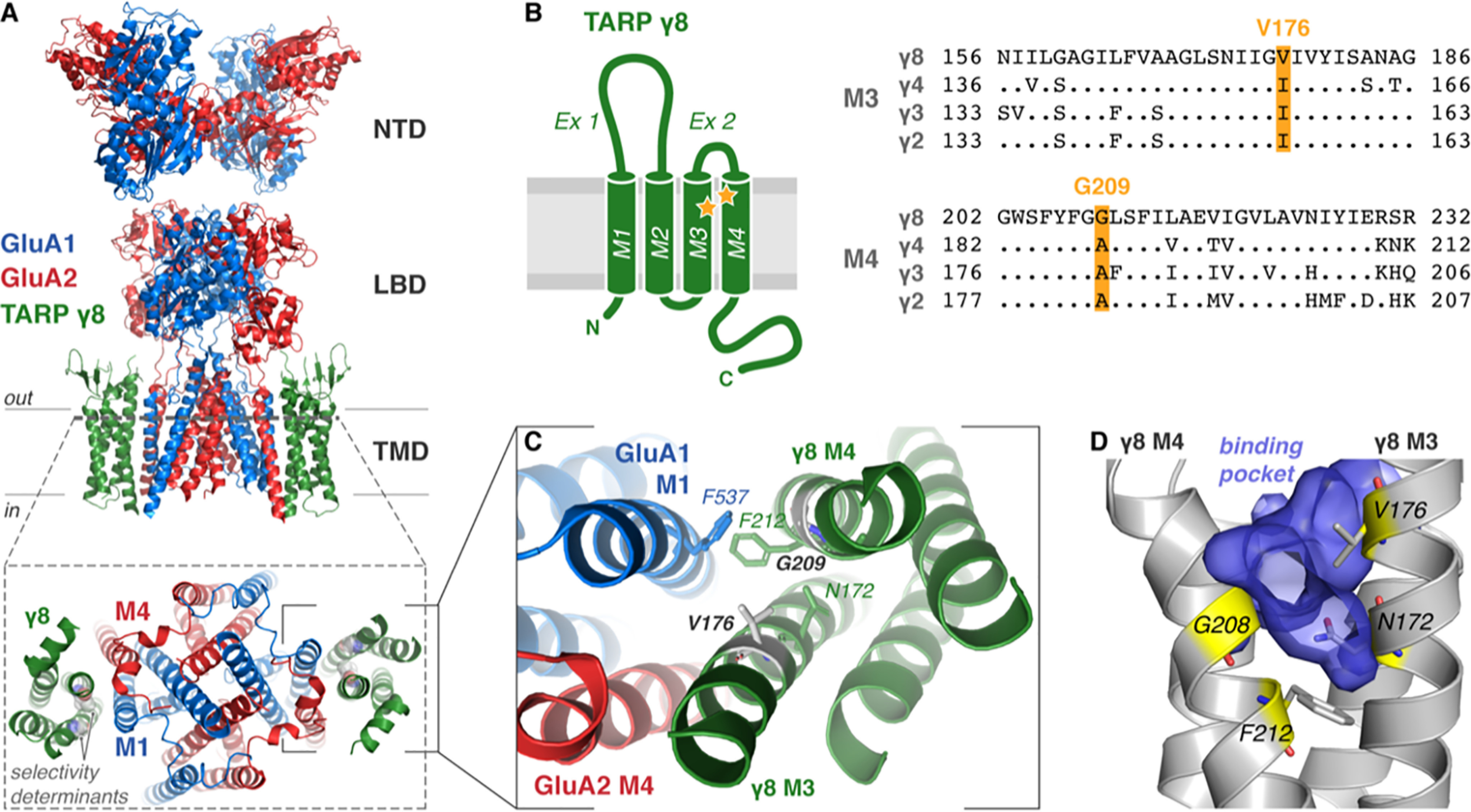
Architecture and proposed ligand-binding pocket of TARP γ8. A, cryo-EM structure of heteromeric GluA1/2 AMPAR (blue/red, respectively) in complex with TARP γ8 (green; PDB 6QKZ) demonstrating the domain architecture (NTD, N-terminal domain; LBD, ligand-binding domain; TMD, transmembrane domain). A top view onto the pore region (bottom, from dotted slice) demonstrates γ8 association with M1 and M4 of the AMPAR. γ8 ligand selectivity-determining residues Gly-209 and Val-176 are shown (gray spheres). B, TARP topology schematic shows the four transmembrane helices (TM1–4) with two extracellular loop elements (Ex1 and Ex2) and both N and C termini located intracellularly. Sequence alignment of type I TARPs (Rattus norvegicus) with divergent residues only shown for TARPS γ2–4 (right) highlights the residues determining γ8 selectivity of ligands (yellow, Val-176 and Gly-209), also marked on the topology map (yellow stars). C, expanded view of the AMPAR-γ8 interaction interface viewed from above. Critical interface residues are depicted as sticks, and selectivity-determining residues are gray. D, the important ligand-binding residues (yellow sticks, see Maher et al. 2016 (10)) line the surface of a binding pocket (blue volume) of an all-atom model of γ8 that forms in MD simulations between TM3 and TM4 of γ8 (gray).
