Figure 8.
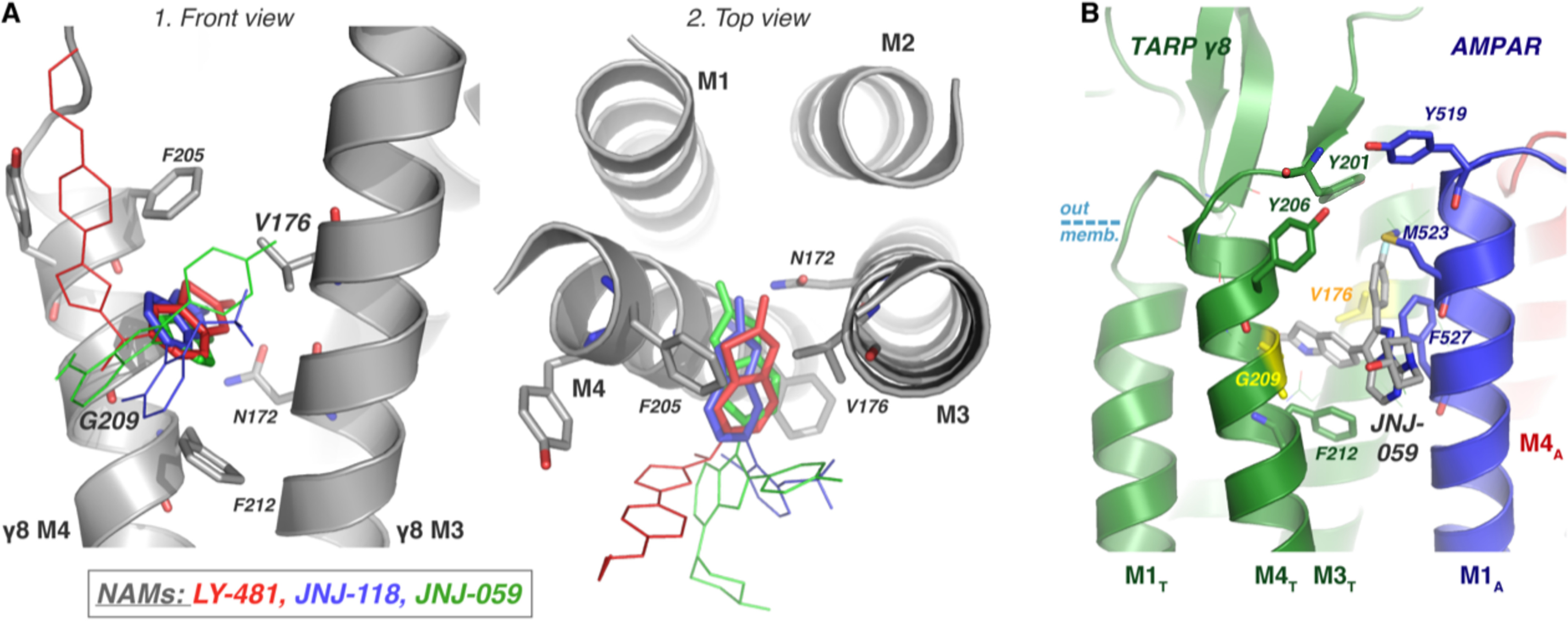
Summary of predicted binding modes. A, the three AMPAR-TARP NAMs (LY-481, red; JNJ-118, blue; JNJ-059, green) all have a similar binding mode in TARP γ8, as depicted on the modeled protein structure, facilitated by γ8 specificity residues Gly-209 and Val-176, and stabilized by Asn-172. The oxindole isostere moiety, conserved between the three ligands, are shown in bold, and the variable regions are depicted as thin lines. B, the pose shown in Fig. 4C1 was placed in the context of the TARP/AMPAR complex (PDB 6QKC) with minor changes. As shown, the ligand predominantly interferes with interactions between M4T and M1A. AMPAR residues Phe-527 and Met-523, which contact TARP in the cryo-EM structure, and TARP tyrosines capping the pocket (Tyr-201 and Tyr-206) may be primary contact points. We note that the exact pose of the variable position is currently unclear.
