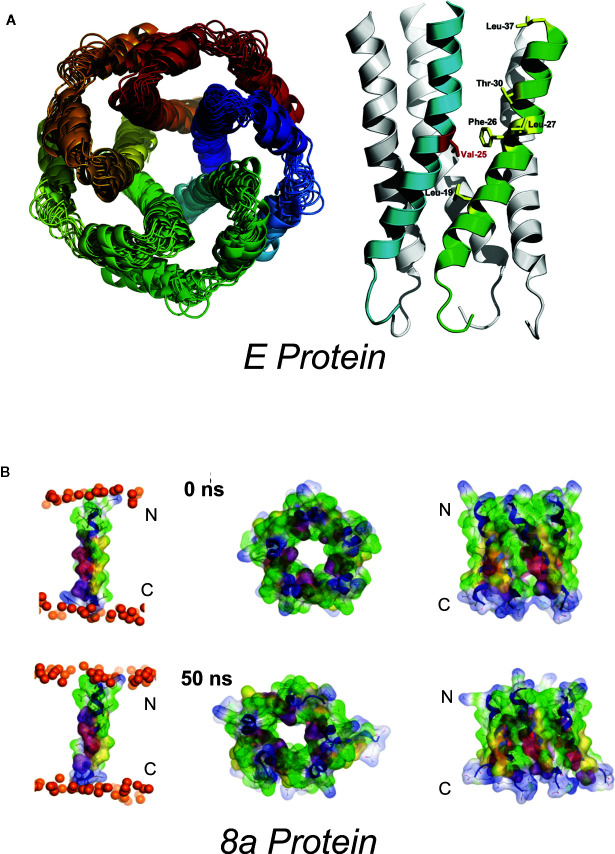
Figure 1.
Proposed SARS-Cov E and 8a structures (A) Proposed homopentameric structure of the E protein (32), viewed (left) through the membrane, and (right) on the plane of the membrane. The structural model includes a ~2Å radius constriction, formed by the sidechains of V25 and V28, which could conceivably act as a channel gate, and an extended central “pore” of <6Å in radius (34) [From Surya et al. BBA-Biomembranes 2018 1860: 1309-1317. With publisher’s permission] (B). Proposed pentameric structure of the 8a protein. (left) The single transmembrane domain (TMD) 8a 1–22 is shown at the beginning (0 ns) and end of a 50-ns MD simulation, (right). Top view (left) and side view (right) of a pentameric bundle of 8a 1–22 at the beginning (upper) and end of 50 ns MD simulation (lower). The protein backbones are drawn in blue with the side chains shown as sticks and van der Waals surface representation. Residues Thr-8, Ser-11, and 214 are shown in pink and light red, respectively. All cysteine residues, Cys-9, 213, and 217, are shown in yellow. Phosphorous atoms of the lipids are shown in orange spheres. Lipid and water molecules are omitted for clarity. [Relabeled from Hsu et al., Proteins. 2015; 83: 300–308. With publisher’s permission].