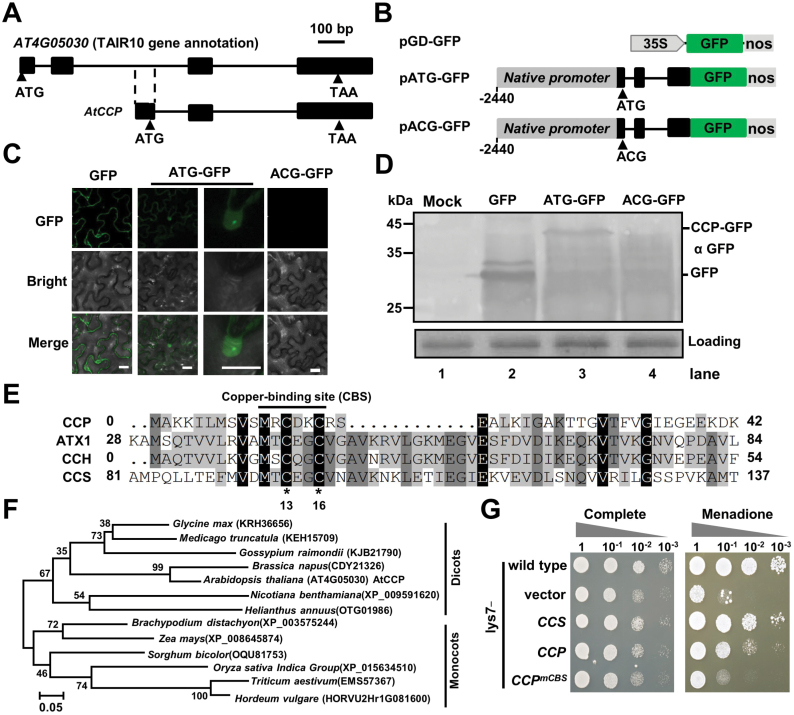
Fig. 1.
Identification of a novel copper chaperone in Arabidopsis. (A) Genome structure of AT4G05030 from TAIR10 gene annotation (upper) or the identified CCP cDNA (bottom). Exons and introns are indicated by black boxes and lines, respectively. The start codon (ATG) and stop codon (TAA) of the predicted small ORF are indicated by arrowheads. (B) Illustration of binary vectors for expressing GFP or GFP-fused proteins in N. benthamiana leaves. In pATG-GFP, the DNA genome fragment between 2440 bp upstream of the CCP start codon to the end of the CCP ORF was amplified from Col-0 and fused to the N-terminus of GFP. In pACG-GFP, the predicted start codon ATG of pATG-GFP was mutated to ACG. (C) Confocal micrographs examining GFP, ATG–GFP, or ACG–GFP expression in agroinfiltrated N. benthamiana leaves at 3 dpi. Scale bar=20 μm. (D) Western blotting analyses showing accumulation of GFP, ATG–GFP, or ACG–GFP in the samples of (C) with anti-GFP antibody. Buffer-infiltrated leaves served as a mock control. The Coomassie brilliant blue staining was used as the protein loading control. (E) Alignments of Arabidopsis CCP, ATX1, CCH, and CCS. The conserved copper-binding site (CBS) is indicated by a line. (F) Phylogenetic trees of plant CCP orthologs based on amino acid sequences. Accession numbers or locus are shown. Scale bar=0.05 amino acid substitutions per site. (G) Complementation of the growth phenotype of the S. cerevisiae lys7 mutant cells transformed with empty plasmid (vector), or plasmids expressing Arabidopsis CCS, CCP, or CCPmCBS. Wild-type S. cerevisiae and complemented lys7 mutant cells were assayed for growth on complete medium or medium with 25 μM menadione. Cells were spotted in 10-fold serial dilutions starting at OD600=0.1, and incubated at 30 °C for 3 d. (This figure is available in color at JXB online.)