FIGURE 1.
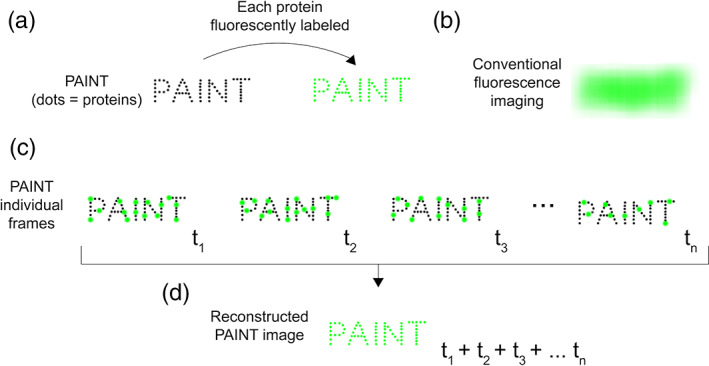
Cartoon illustration of the principle of the PAINT method of SMLM. PAINT achieves super‐resolution by summing sparse, temporally‐separated localization events. (a) A biomolecular structure “PAINT” (dimensions of the order of 500 × 2000 nm) composed of multiple proteins. Individual proteins are shown as black dots. If each protein is directly fused to a fluorescent molecule (green dots), (b) conventional fluorescence imaging cannot resolve individual fluorophores, so the PAINT structure is fluorescent, but individual proteins cannot be visualized—because the proteins are too close together to be resolved by diffraction‐limited microscopy. (c) SMLM by PAINT. The proteins in the biomolecular structure are not directly fused to a fluorescent molecule. They are only visible when a fluorescent molecule transiently binds to any of them, resulting in intense bursts of fluorescence (green spots). Such data are collected iteratively over time (t 1, t 2, t 3, … t n). At each timepoint, a different subset of the proteins is bound to the fluorescent molecule. (d) The localization events collected at each timepoint (t 1, t 2, t 3, … t n) in panel (c) are summed to generate a final super‐resolution image, in which the location of each protein can now be resolved
