FIGURE 3.
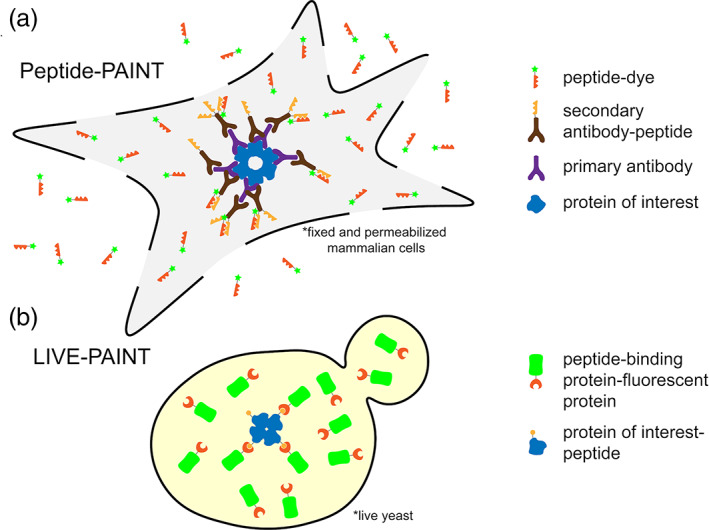
Cartoon representations of using Peptide‐PAINT in fixed, permeabilized mammalian cells (a) and LIVE‐PAINT inside live yeast (b). (a) The protein of interest (blue) is bound by a primary antibody (purple). A secondary antibody (brown), which binds to the primary antibody, is attached to one or more peptides (orange sawtooth). A peptide (red sawtooth) that interacts with the antibody‐linked peptide is synthesized with a fluorescent dye (green star) attached. Cells are fixed and permeabilized and the peptide‐dye fusion (red sawtooth‐green star) is added exogenously and can diffuse in and out of the cell. Excess antibodies and fluorescently labeled peptide can be washed out prior to imaging in TIRF, which further decrease the background. (b) The protein of interest (blue) is fused to a peptide (orange circle), at the gene level, and integrated into the chromosome. A peptide‐binding protein, comprising the recognition element for the peptide (red crescent) is fused to a fluorescent protein (green barrel), at the gene level, and integrated into the chromosome. Labeling is performed inside live cells, with the expression level of the labeling protein controlled. Background from unbound labeling protein is reduced by data acquisition in TIRF
