Fig. 7.
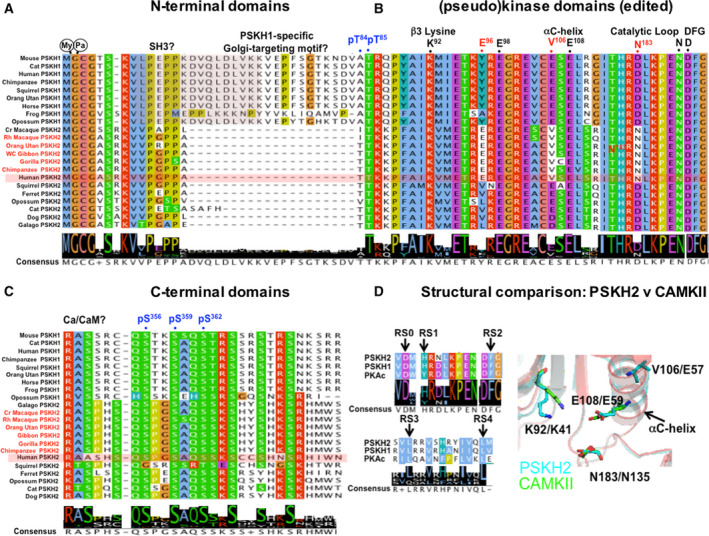
Comparative analysis of vertebrate PSKH1 and PSKH2. Amino acid alignment of selective conserved and unique regulatory sites and catalytic motifs (above, black) comparing human and other primate PSKH2 orthologues highlighted (red) with PSKH1 and nonprimate PSKH2. (A) PSKH1 and PSKH2 N termini from 22 vertebrate genomes aligned in Jalview 88, with human PSKH2 shaded. (B) Selected catalytic amino acids/motifs aligned (conserved primate PSKH2 changes in red, human numbering), with potential human PSKH2 phosphorylation sites shown in blue. Primate‐specific hot spots of amino acid identity in PSKH2 orthologues are evident. (C) Divergent PSKH1 and PSKH2 C termini, with speculative human phosphorylation sites shown in blue. (D) Five classical putative RS residues found in PKA catalytic domain 89 are also conserved in PSKH1 and PSKH2 (arrows), suggesting that an ‘active‐like’ fold is possible in both. PSKH2 (cyan sticks) exhibits catalytic potential based upon HHPred model using CAMKII (green sticks), the closest kinase at the amino acid level for which a structure is available. The predicted PSKH2 αC‐helix catalytic residue Glu108 might interact with the β3 Lys92, equivalent to the Glu59:Lys41 interaction formed in active Ser/Thr kinases. Val106 (notably a Glu side chain in nonprimate PSKH2) also lies on the αC‐helix. Asn183 of PSKH2 adopts a similar conformation to CAMKII Asp135Asn (note that this mutation was introduced experimentally in order to generate a kinase‐inactive mutant for crystallography). This analysis raises the possibility that N183D PSKH2, perhaps alongside other PSKH2‐specific residues found at Glu96 and Val106, might promote catalytic activity in the presence of Ca2+/calmodulin.
