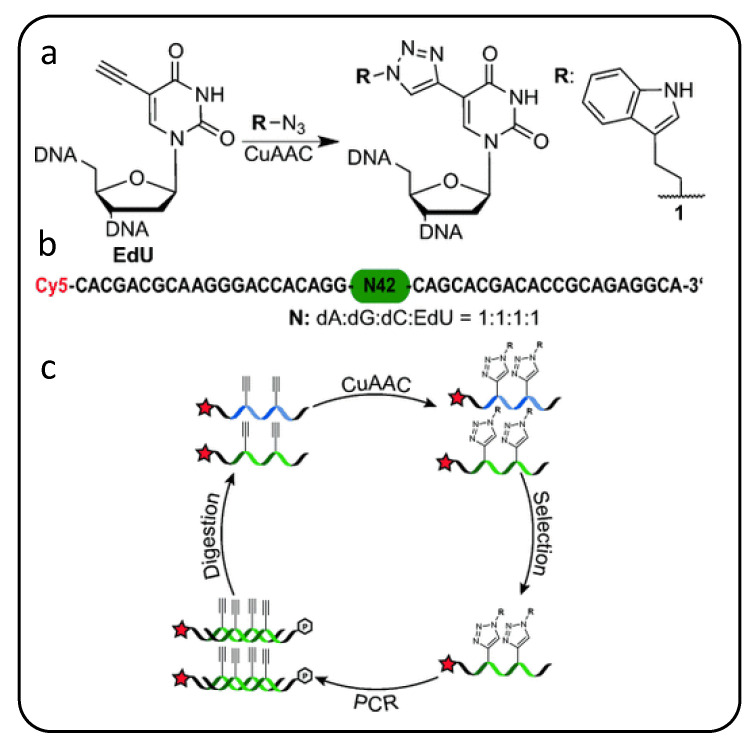
Figure 3.
Schematic illustration of modified sequence libraries incorporating synthetic handles to side-step PCR amplification issues during click-SELEX. (a) To chemically modify nucleotides, C5-ethynyl handle-modified uracil nucleobases in the precursor sequences are converted to chemical groups (shown as R in 1) using click chemistry to prepare (b) random sequence library. (c) During click-SELEX a modified library is incubated with a target (not shown). Following selection and recovery of target-binding sequences, the modified library must be “reverse transcribed” back to the precursor sequences bearing only the C5-ethynyl handle to facilitate PCR amplification-based enrichment of aptamer candidates. Finally, the now enriched candidate sequence population is subjected to a click reaction once more to reintroduce the chemical modification and begin the next selection cycle. (Reprinted with permission from Tolle et al. [89] Copyright 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim).