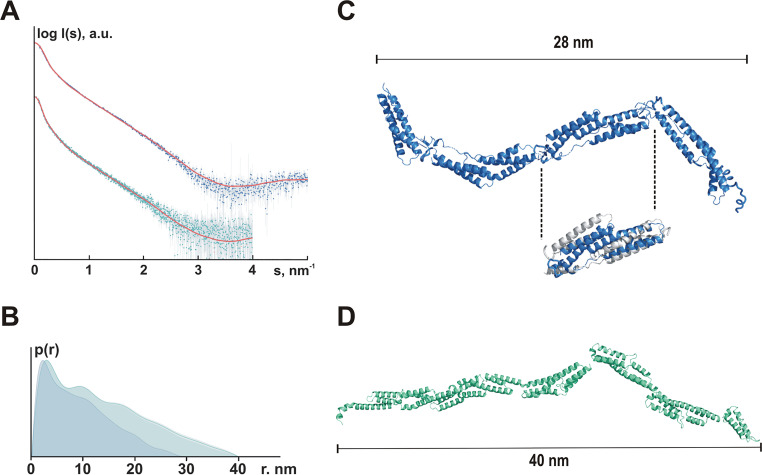
FIG 6.
Organization of F- and FG-repeats in solution. (A) Experimental SAXS data from the Embp F-repeats construct (blue dots with error bars) and the Embp FG-repeat construct (cyan dots with error bars) fitted (red lines) with scattering computed from the rigid body models shown in Fig. S4A and B, respectively. For the F-repeats, the radius of gyration Rg estimated from Guinier approximation was 7.0 ± 0.2 nm, and the maximum dimension Dmax was 28 nm; for the FG-repeats, the Rg was 10.7 ± 0.5 nm, and the Dmax was 40 nm. (B) Pair distance distribution functions computed from the F-repeats (blue, Dmax = 28 nm) and the FG-repeats (cyan, Dmax = 40 nm) SAXS data. (C) Rigid body model of four F-repeats. Despite using several modeling approaches, it was not possible to obtain a satisfactory fit at higher angles (2 to 5 nm−1), although the overall shape resembled the respective ab initio model. Assuming that the structure of the individual F-repeats in solution was different from that in the crystal, the F-repeat structure was refined to fit the higher angle scattering data using the program SREFLEX (69). The RMSD between the original structure and the refined model (cutout: gray, F-repeat according to crystal structure analysis; blue, SREFLEX-refined F-repeat model) was 0.57 nm. RANCH (31) was then used to generate a model consisting of four repeats of the refined structure (best fit, χ2 = 2.24). Fits were computed by CRYSOL (68). (D) Rigid body model of six FG-repeats. RANCH (31) was used to generate a model consisting of six FG-repeats (best fit, χ2 = 1.07). Fits were computed by CRYSOL (68). For the rigid body model, no refinement of the 6GV5 model was required, suggesting that the crystal structure of the domain is preserved in solution.