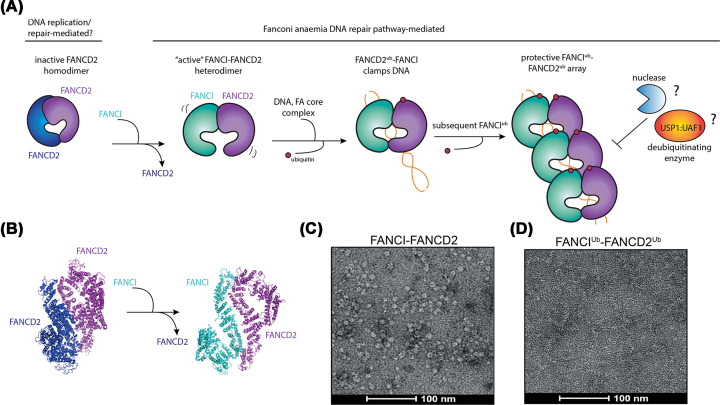
Figure 3. Proposed activation mechanism of FANCIUb–FANCD2Ub filamentous array.
(A) FANCD2 forms an inactive homodimer which is inaccessible to DNA binding. FANCI displaces one of the FANCD2 and assembles into the FANCI–FANCD2 heterodimer, which is the substrate for FA core complex E3 ligase and DNA binding. FANCD2 ubiquitination clamps FANCI–FANCD2 heterodimer on DNA, which further stimulates ubiquitination of FANCI to form protective array against DNA degradation by nucleases or deubiquitination by USP1:UAF1. (B) Comparison of FANCD2 homodimer (PDB ID: 6TNI) and apo-FANCI–FANCD2 heterodimer (PDB: 6VAD). (C) Negative-stained electron micrograph of apo-FANCI–FANCD2. (D) Negative-stained electron micrograph of di-ubiquitinated FANCI–FANCD2 filamentous arrays. FANCD2 is colored in blue or violet, FANCI in cyan, ubiquitin in red and DNA in orange.