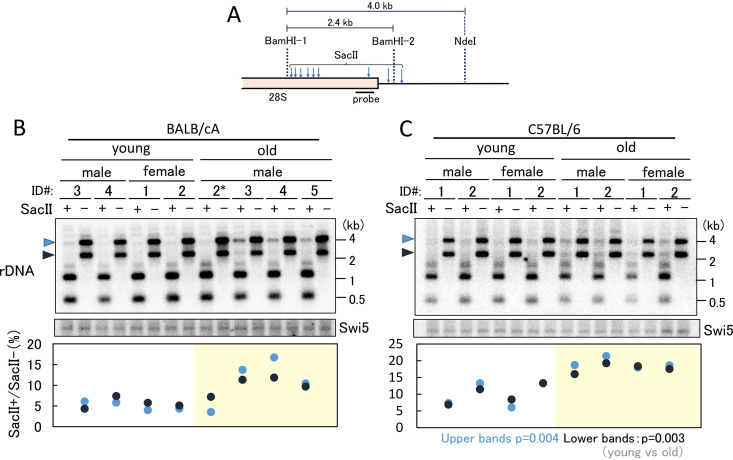
FIG 6.
rDNA methylation in old and young bone marrow cells. rDNA methylation was detected by digestion with the methylation-sensitive enzyme SacII. (A) Position of the probe for Southern blot analysis in panels B and C and recognition sites for BamHI and SacII. (B and C) Ratio of copy numbers of methylated rDNA in young and old mice. Southern blot analysis was used to detect the ratio in undigested bands by SacII (top). The positions of undigested bands are indicated by arrowheads on the left-hand side of the panels. The SWI5 gene served as a loading control (middle). A single-copy gene, SWI5, was detected on the same filter used for the experiment shown in the upper panel. Analysis of rDNA that failed to digest with SacII was performed (bottom). The signal intensities of the undigested rDNA (SacII+) and nondigested (SacII−) bands were measured, and the ratios were plotted. The black and blue dots show the results corresponding to the bands indicated by the black and blue arrowheads, respectively, in the top panel. Identification (ID) numbers of individual mice that were used to isolate the bone marrow cells are given (Fig. 3). P values were calculated from the average of the values for young and old mice.