Figure 4.
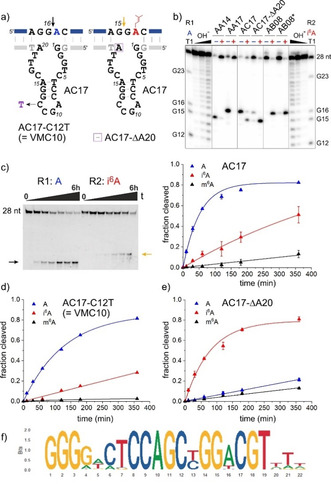
a) Predicted secondary structure of AC17 shown to cut unmodified RNA at G16 and i6A‐RNA at G15. Sites of important mutations are indicated. b) PAGE analysis of cleavage sites of AC17 in comparison to AA14, AA17 and AB08. Note that this analysis was performed with 5′‐32P‐labelled RNA (in contrast to the 3′‐fluorescein‐labelled RNAs used in Figures 2 b and 3 b). c) PAGE analysis with 3′‐fluorescein‐labelled RNAs and kinetic graphs for cleavage of R1 (A), R2 (i6A) and R3 (m6A) RNA). d,e) kinetic graphs for cleavage of R1 (A), R2 (i6A) and R3 (m6A) RNA with VMC10 (d) and AC17‐ΔA20 (e). f) Sequence logo for alignment of 45 AC17 variants with >50 reads and maximal 4 mutations.
