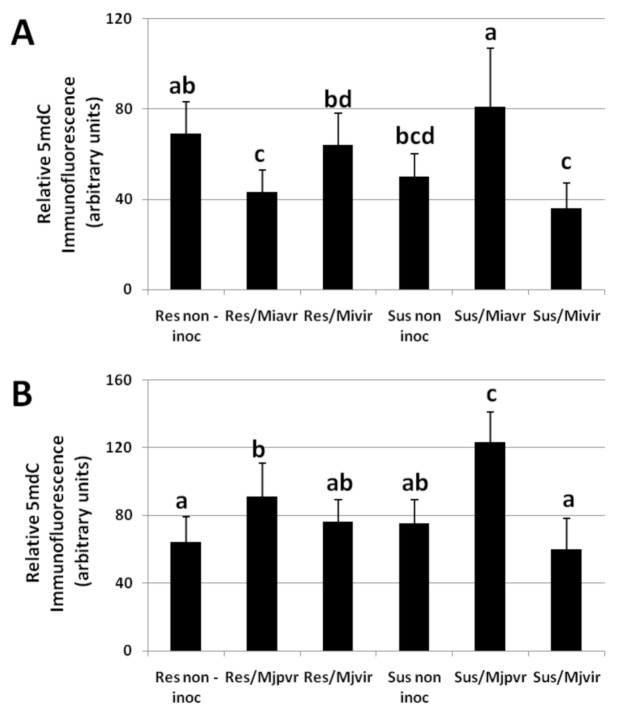
Figure 4.
Arbitrary units of methylation on total DNA in roots of susceptible and resistant tomato plants un-inoculated or inoculated with 2 couples of field populations/virulent isolates of RKNs. Data were taken at 7 DAI. DNA was extracted from resistant (Res non-inoc, Rossol) and susceptible (Sus non-inoc, Roma VF) un-inoculated plants. In A, DNA was also extracted from roots of Rossol and Roma VF plants inoculated with the avirulent Meloidogyne incognita field population Mi-Vfield and the selected virulent isolate SM2V: Rossol/Mi-Vfield (Res-Miavr); Rossol/SM2V (Res-Mivir); Roma VF/Mi-Vfield (Sus-Miavr); Roma VF/SM2V (Sus-Mivir). In B, DNA was also extracted from roots of Rossol and Roma VF plants inoculated with the partially virulent Meloidogyne javanica field population Mj-Tunc2field and the selected virulent isolateSM11C2: Rossol/Mj-TunC2field (Res-Mjpvr); Rossol/SM11C2 (Res-Mjvir); Roma VF/Mj-TunC2field (Sus-Mjpvr); Roma VF/SM11C2 (Sus-Mjvir). Values are expressed as arbitrary units of relative 5mdC immunofluorescence and as means (n = 6) ± SD. Means were separated by a Duncan’s Test (Significance Level: 0.05).