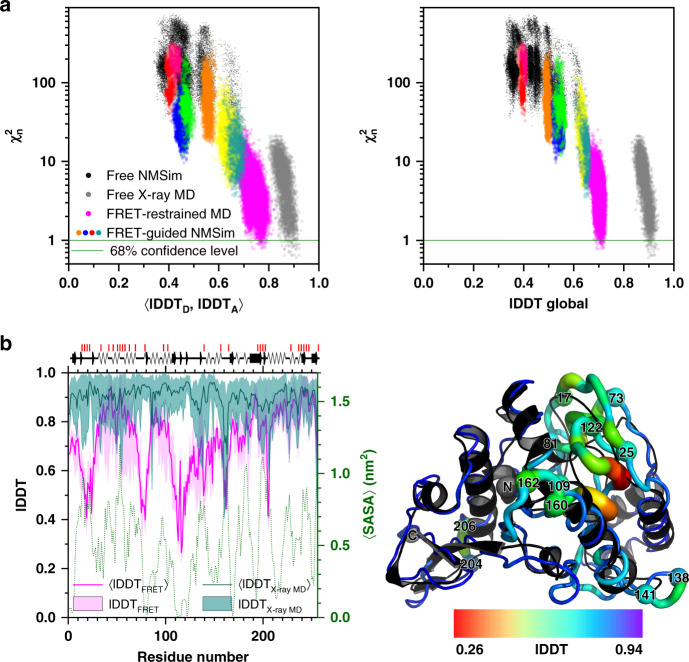
Fig. 5. Local accuracy and uncertainty of FRET-assisted structure models of the E. coli protein YaaA.
a The left panel displays FRET values and local distance difference test (lDDT) scores against the crystal structure (target) for different conformations (points). 〈lDDTD, lDDTA〉 stands for the average over lDDT scores for residues that were in silico labeled by donor (D) and acceptor (A) fluorophores. Black points stand for unrestrained NMSim sampling starting from homology models. Colored points represent FRET-guided NMSim simulations. Magenta points represent FRET-restrained MD simulations. Guided simulations stemming from different homology models are shown in different colors. Gray points represent unrestrained MD simulations, started from the crystal structure of the YaaA protein, for reference. In the right panel, the x axis shows global lDDT scores: average number of conserved distances in the structure over four tolerance thresholds: 0.5, 1, 2, and 4 Å38. The lDDT scores were computed using only distances between α carbon atoms. b The magenta line represents ensemble-average lDDT score for FRET-selected conformers (〈lDDTFRET〉). The shaded magenta area represents the range of lDDT scores in the FRET-selected ensemble (lDDTFRET). The cyan line represents ensemble-average lDDT for conformers from 500 ns of unrestrained MD simulations started from the crystal structure (〈lDDTXray MD〉), the shaded area indicates the range of lDDT scores in this simulation (lDDTX-ray MD). The green dotted line shows the solvent-accessible surface area averaged over 500 ns of unrestrained MD simulations and smoothed over the window of five residues. Secondary structure elements of the crystal structure are shown above the graph. Red vertical lines on top of the secondary structure information indicate the labeling positions. The putty plot shows the FRET-selected conformer; the color and thickness of the tube represent the residue-wise lDDT against the crystal structure (black cartoon), red thick regions correspond to less accurately predicted residues. Data are provided in the Source Data file.