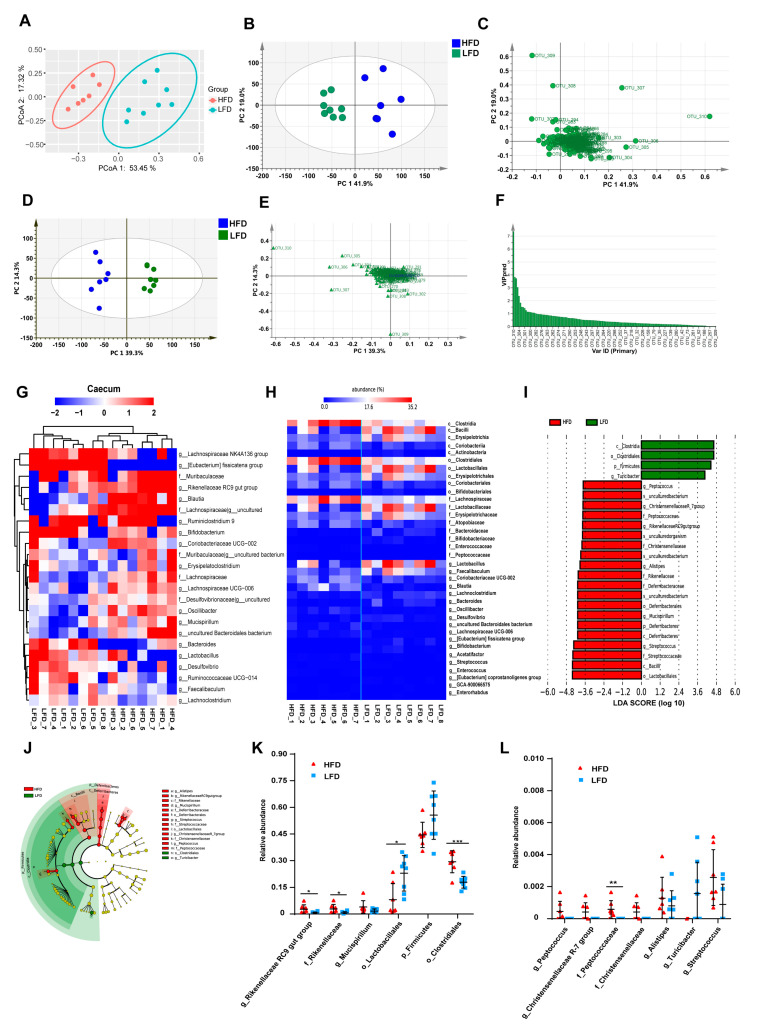
Figure 4.
Different diets resulted in different gut microbial clusters and different dominant bacteria in the cecum. (A) Principal coordinate analysis (PCoA) of gut microbiota for the HFD and the LFD groups (PCoA 1 = 53.5%, PCoA 2 = 17.3%). (B) Principal component analysis (PCA) score scatter plots for gut microbiota for the HFD and the LFD (PC 1 = 41.9%, PC 2 = 19.0%). (C) PCA loading scatter plot of gut microbiota for the HFD and the LFD groups. (D) The orthogonal partial least square-discriminate analysis (OPLS-DA) score scatter-plot of gut microbiota for the HFD and the LFD groups (PC 1 = 39.3%, PC 2 = 14.3%). (E) The OPLS-DA loading scatter plot of gut microbiota for the HFD and the LFD groups. (F) Values of variable importance for the projection (VIP) for the predictive components (VIP predictive) of gut microbiota. (G) Heatmap of the proportion of the 23 operational taxonomic units (OTUs) determined as dominant bacteria, with rows clustered by microbiota similarity according to the Euclidean distance, and columns clustered by operational taxonomic units (OTUs) that occur more often together. (H) Heatmap of the statistical analysis of taxonomic and functional profiles (STAMP) showed significant differences in the microbiota of the HFD and the LFD groups. (I) Linear discriminant analysis (LDA) scores of gut microbiota for the HFD and the LFD groups. (J) Cladograms representing the LDA effect size (LEfSe) results for the HFD and the LFD groups. (K,L) Dot plots representing the proportional abundance of OTUs from LEfSe results. Mean values ± S.D. are plotted. Asterisks indicate significant differences (unpaired two-tailed Student’s t-test, * p < 0.05, ** p < 0.01, *** p < 0.001, n = 7–8 mice per group).