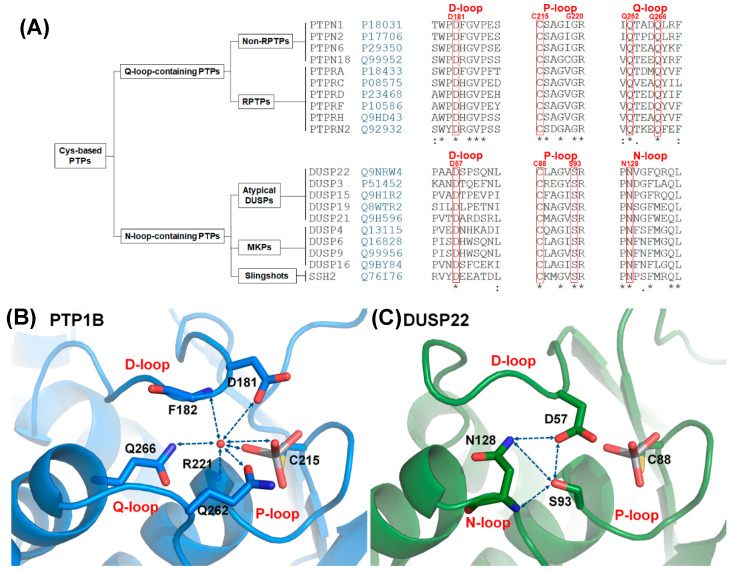
Figure 1.
The D-, P-, N-triloop (DPN–triloop) interaction in N-loop-containing protein tyrosine phosphatases (PTPs). (A) The sequence alignment showed the difference between Q-loop-containing PTPs and N-loop-containing PTPs. N-loop-containing PTPs had an N-loop motif (PNXXF), which was different from the Q-loop motif (QXXXQ). The key residues (red) in PTP1B and DUSP22 are labeled above the sequence alignment. (B) In the structure of PTP1B (PDB ID: 3I80), structural water (red ball) can form the hydrogen bonding with the side chain of D181, the side chain of Q262, the side chain of Q266, the backbone amide of F182, the backbone amide of R221, and the oxygens of the ligand in active site. This water-mediated DPQ-triloop interaction connected the D-loop, the P-loop, the Q-loop, and the ligand, and aligned these residues to the correct position. (C) In the structure of DUSP22 (PDB ID: 6LVQ), the active site contained a hydrogen bonding network named the DPN–triloop interaction, which was formed by three residues: D57 in the D-loop, S93 in the P-loop, and N128 in the N-loop. The side chains of D57, S93, and N128 formed hydrogen bonds with each other, and the backbone amide of N128 formed hydrogen bonding with the side chain of S93. Hydrogen bonding is displayed as blue dotted lines, and the residues as well as the ligand, VO4, are shown by sticks.