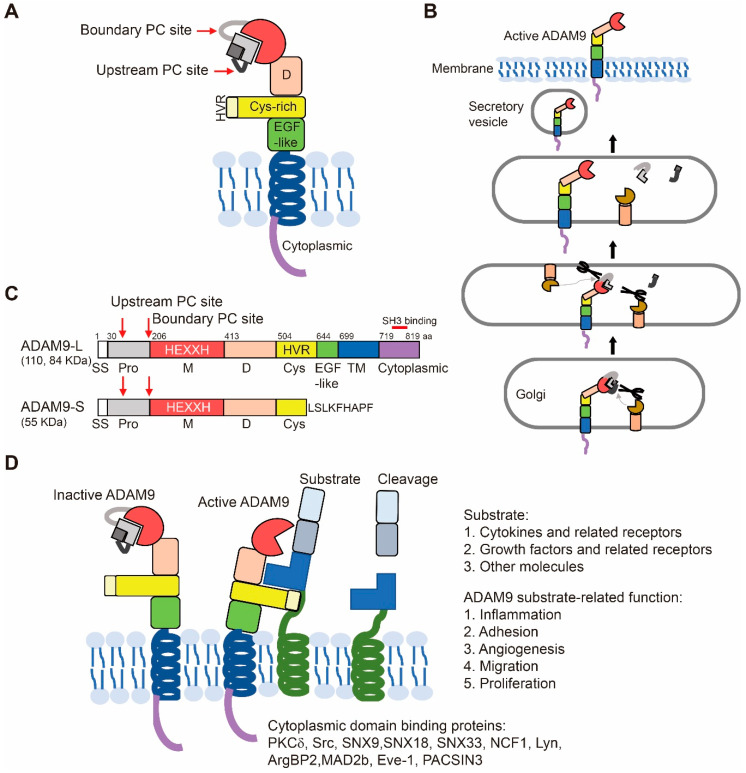
Figure 1.
Characterization of a disintegrin and metalloprotease 9 (ADAM 9). (A) The organization of the C-shape structure when anchored to the membrane is shown. (B) ADAM9 is activated during transport via Golgi bodies to the cell membrane by removal of the inhibitory pro-domain. The pro-domain cleavage is performed at the two PC sites by furin or proprotein convertase. (C) Schematic diagrams of the domain structure of the long (ADAM9-L, 110 and 84 KDa) and short (ADAM9-S, 55 KDa) forms of ADAM9. ADAM9-S lacks exon 12 from the ADAM9-L genetic sequence. aa, amino acid. SS, signal sequence. Pro, pro-domain. M, metalloprotease domain. D, disintegrin domain. Cys, cysteine-rich. TM, transmembrane. PC site, proprotein convertase (PC) consensus cleavage site. HVR, hyper-variable region. (D) Active ADAM9 recognizes its substrates via the HVR of the cysteine-rich domain and releases the extracellular fragments of membrane-bound cytokines and growth factors. It also cleaves receptors and other molecules for signal transduction.