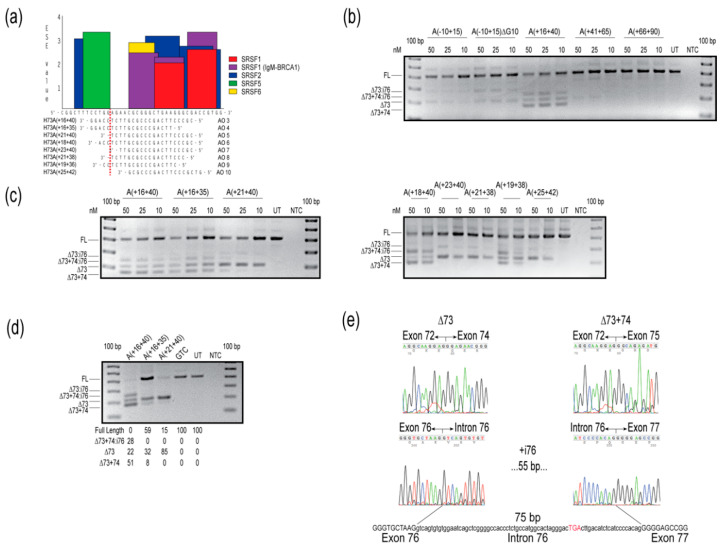
Figure 2.
Evaluation of splice-switching antisense oligomers (AOs) targeting COL7A1 exon 73 in healthy human fibroblasts. (a) Locations of AOs targeted to remove COL7A1 exon 73, designed by micro-walking across the COL7A1 H73(+16+40) annealing site, and the exon splice enhancer (ESE) motifs, predicted by ESEfinder 3.0 [12]. Relative splice factor binding scores, as predicted by ESEfinder 3.0 are indicated on the y-axis. The color code indicates the putative binding sites for serine and arginine rich splicing factors (SRSFs). (b) Reverse transcription polymerase chain reaction (RT-PCR) analysis of COL7A1 transcripts after transfection with 2′-O-methyl modified bases on a phosphorothioate backbone (2′-OMe) AOs, at concentrations indicated above the gel image. (c) Reverse transcription PCR analysis of COL7A1 transcripts after transfection with overlapping 2′-OMe AOs, at concentrations indicated above the gel image. (d) Reverse transcription PCR analysis of COL7A1 transcripts after nucleofection with PMOs at a concentration of 150 µM in the cuvette, with relative abundance of amplicons (as percentage) shown below the gel image. Data are representative of at least two independent experiments. (e) Sanger sequencing data identifies the different amplicons resulting from AO treatment. Intronic sequence is represented in lower case, exonic sequence as upper case text. The premature termination codon in intron 76 is indicated by red upper case text. The gels were cropped for presentation. Full gel images are presented in Figure S2. GTC, Gene Tools control; NTC, no template control; UT, untreated; bp, base pairs; FL, full-length amplicon; nM, nanomolar; i, intron.