Figure 3. Disruption of LaminA/C decreases nuclear stiffness and changes structure.
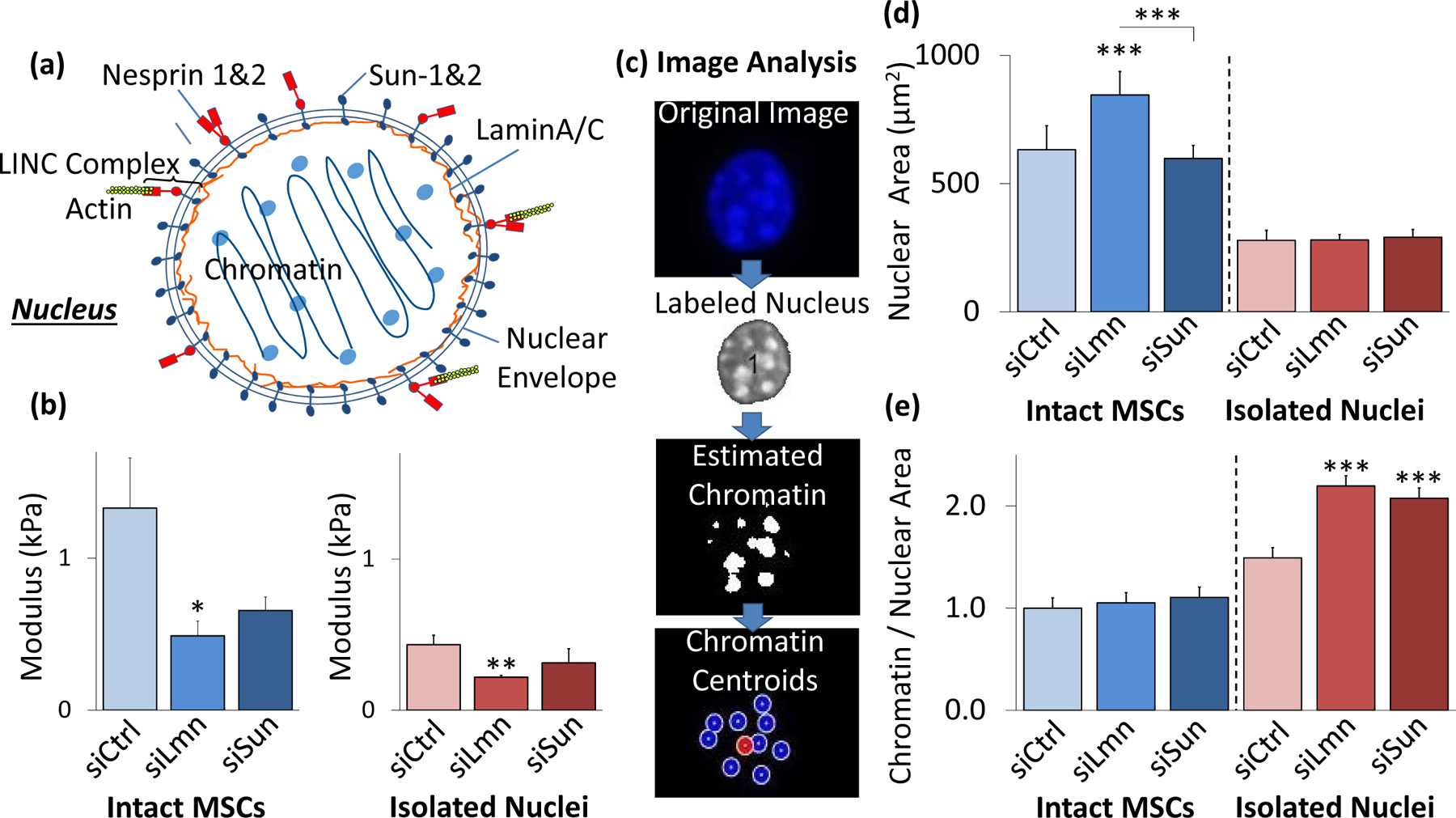
a) Schematic representation of the nucleus with nucleoskeletal and cytoskeletal connection components illustrated, including LaminA/C, Sun-1&2, Nesprin, and the KASH domain. b) SiRNA against nuclear LaminA/C (siLmn) and Sun-1&2 (siSun) decreased both isolated nuclear and intact MSC stiffness. Compared to siCtrl (N=21), siLmn and siSun treatments showed a significant decrease in the intact MSC modulus by 63.2% (p<.05, N=22) and 50.7% (p>.05, N=10), respectively. Likewise, when compared to siCtrl (N=21) the elastic modulus of isolated nuclei decreased by 49.7% (p<.01, N=21) and 27.7% (p>.05 N=10) in siLmn and siSun groups, respectively. c) A MATLAB code was constructed to evaluate differences in nuclear area and nucleoli size as determined by live Hoechst 33342 staining (see Supplementary Methods). d) Nuclear staining via Hoechst 33342 and epifluorescence imaging revealed that nuclear area increased by 33% within siLmn treated intact MSCs (p<.001, N=73), but not under Sun-1&2 depletion when compared to siCtrl (N=104). Nuclear area showed no significant changes within isolated nuclei control or experimental groups (N=245). e) Chromatin area to nuclear area ratios were calculated for all MSC and isolated nuclei for siCtrl, siLmn and siSun groups. Compared to controls (N=806), intact MSC did not show differences in chromatin to nuclear area ratio for LaminA/C (N=647) andSun-1&2 MSCs (N=489). In contrast, isolated nuclei showed significant increases in chromatin to nuclear area ratios for both LaminA/C (47.2%, p<.001, N=502) and Sun-1&2 (39.1%, p<.001, N=612) depleted nuclei compared to controls (N=416). * p<.05, ** p<.01, *** p<.001, against control or against each other.
