Figure 5. Isolated nuclei maintain a smaller chromatin-to-nuclear area ratio after vibration.
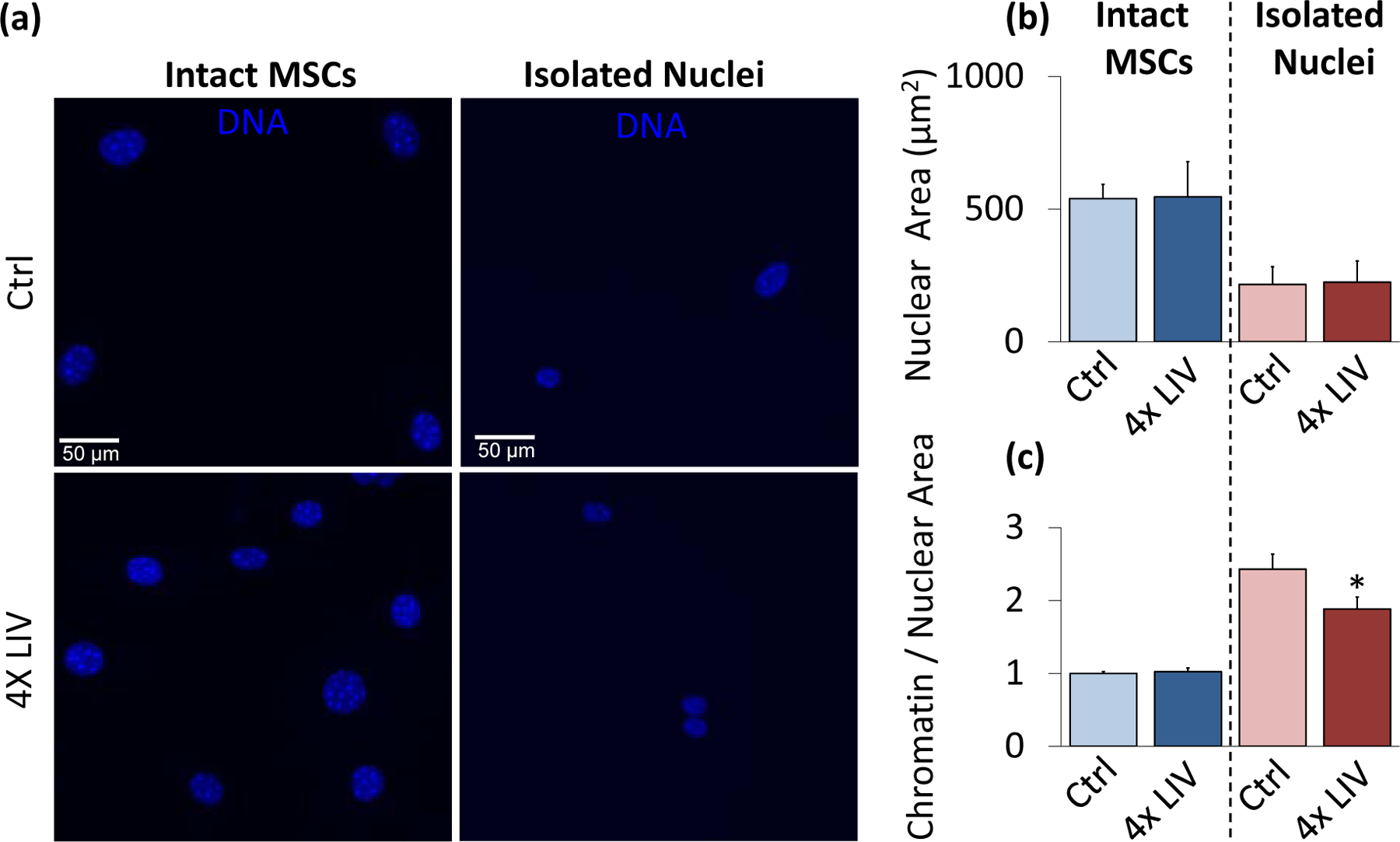
a) Epi-fluorescence images of intact control MSC nuclei (upper left), intact 4x LIV MSC nuclei (lower left), isolated control nuclei (upper right), and isolated 4x LIV nuclei (lower right). Cells were treated with Hoechst 33342 to stain heterochromatin, fixed in 2% paraformaldehyde, and kept in PBS for imaging. b) Nuclear area was measured using MATLAB code to identify nuclear bounds under Hoechst 33342 staining. Intact MSCs subject to 4x LIV (N=216) showed no difference in nuclear area compared to controls (N=214). Likewise, there was no difference in nuclear area between control (N=90) and 4x LIV isolated nuclei (N=102). c) Chromatin measurements were analyzed via MATLAB analysis. Individual chromatin were averaged and compared to respective nuclear area. Intact MSCs subject to 4x LIV (N=2136) showed no difference in chromatin to nuclear area ratio compared to controls (N=2082). Isolated nuclei subject to 4x LIV showed a 25.4% lower chromatin to nuclear area ratio (N=480) than isolated controls (p<.05, N=360). * p<.05, ** p<.01, *** p<.001, against control.
