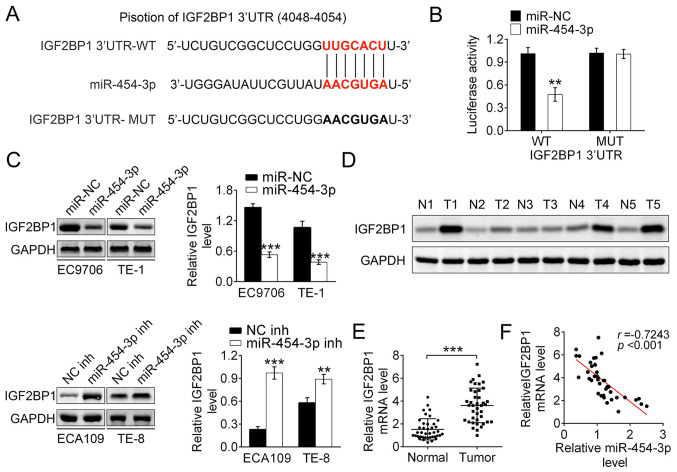
Figure 4.
IGF2BP1 is the direct target of miR-454-3p. (A) The sequences of miR-454 binding sites complemented with the IGF2BP1 3′-UTR (base position of 4048–4054). IGF2BP1 3′-UTR-WT represents the entire 3′UTR sequences of WT IGF2BP1, IGF2BP1 3′-UTR-MUT represents the binding sites were mutated. (B) Luciferase reporter assays were performed in 293T cells to detect the binding affinity of miR-454-3p to the IGF2BP1 3′UTR-WT or MUT. (C) Western blotting was used to analyze how the overexpression or inhibition of miR-454-3p in EC9706, TE-1 ECA109 and TE-8 cells affected the IGF2BP1 protein levels. (D) Western blotting revealed the expression differences of IGF2BP1 protein levels between tumor tissues and normal adjacent tissues in five patients. (E) Reverse transcription-quantitative PCR demonstrated the relative IGF2BP1 mRNA expression levels in tumor tissues or normal adjacent tissues from patients with ESCA (n=40). (F) Pearson correlation test was used to evaluate the correlation between mRNA levels of IGF2BP1 and miR-454-3p expression in tumor tissues from 40 patients with esophageal cancer (r=−0.7243; P<0.001). Results are presented as mean ± SD. **P<0.01 and ***P<0.001. miR, microRNA; IGF2BP1, insulin-like growth factor 2 mRNA-binding protein 1; UTR, untranslated region; WT, wild-type; MUT, mutant; NC, negative control; inh, inhibitor.