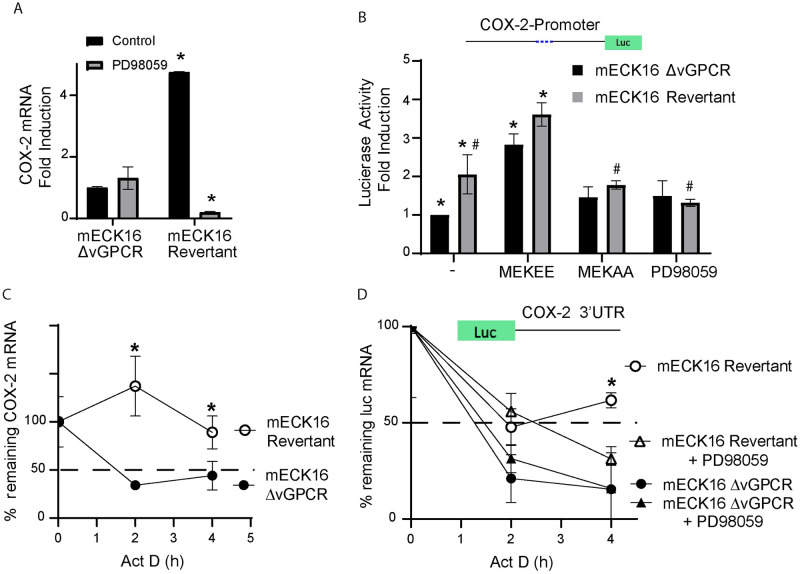
Fig 4. vGPCR regulates COX-2 promoter activity and mRNA stability via ERK1/2 in full KSHV genome bearing cells.
A) Fold-changes in mRNA expression determined by RT-qPCR in mECK16 derived cells (Δ-vGPCR or revertant virus) after treatment with ERK1/2 MAPK inhibitor PD98059 (20uM). COX-2 mRNA was measured in triplicate and is presented as means ± SD. (*P<0.05). B) mECK16 derived Δ-vGPCR and revertant cells were transfected with a luciferase reporter plasmid under the control of the COX-2 promoter (as in Fig 2A). Luciferase activity is expressed as fold induction relative to control cells. Cells were co-transfected with plasmids expressing constitutively active and dominant negative MAP kinase kinases (MEKEE and MEKAA respectively) or treated with the MEK/ERK1-2 inhibitor PD98059. (*) Indicates significant differences from mECK16 Δ-vGPCR untreated cells (P<0.05). (#) Indicates significant differences between mECK16 revertant untreated cells (P<0.05). C) mRNA stability assay in mECK16 derived (Δ-vGPCR and revertant) cells. Actinomycin D (5 μg/ml) was added (t = 0) to arrest transcription, and mRNA levels of COX-2 were analyzed by qRT-PCR following a time course (4 hours). COX-2 mRNA was measured in triplicate and is presented as means ± SD. (*P<0.05). D) mRNA stability assay in mECK16 derived (Δ-vGPCR and revertant) cells transfected with a reporter plasmid containing the COX-2 3’UTR region cloned downstream of the luciferase ORF in the presence or absence of the MEK/ERK1-2 inhibitor PD98059 (20 uM). Actinomycin D (5 μg/ml) was added (t = 0) to arrest transcription, and mRNA levels of Luciferase were analyzed by qRT-PCR following a time course (4 hours). Luciferase mRNA was measured in triplicate and is presented as means ± SD. (*P<0.05).