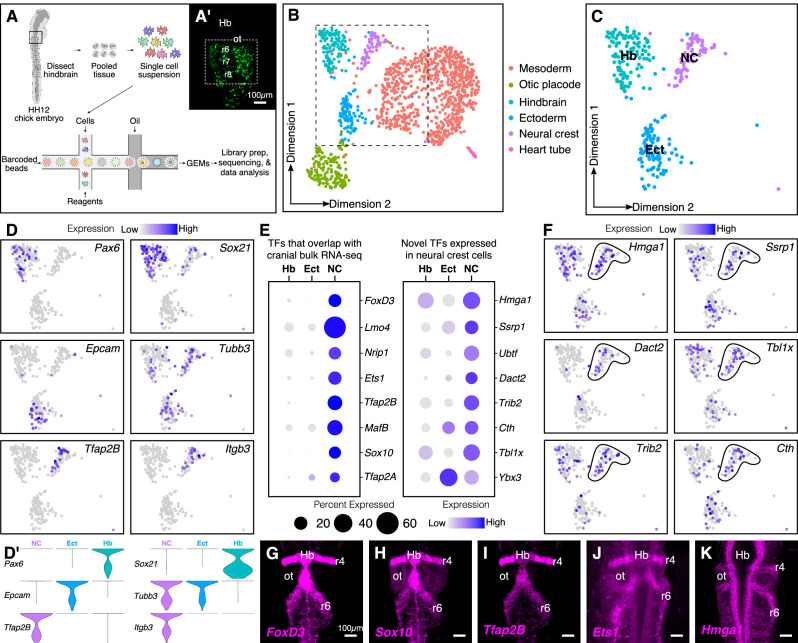
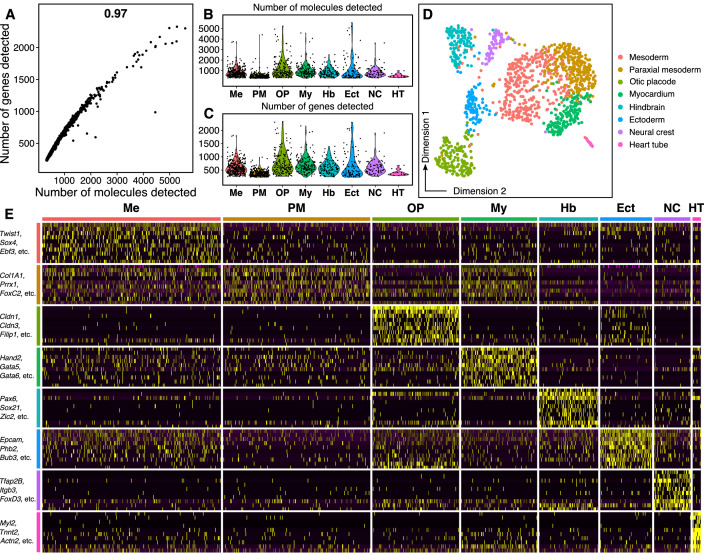
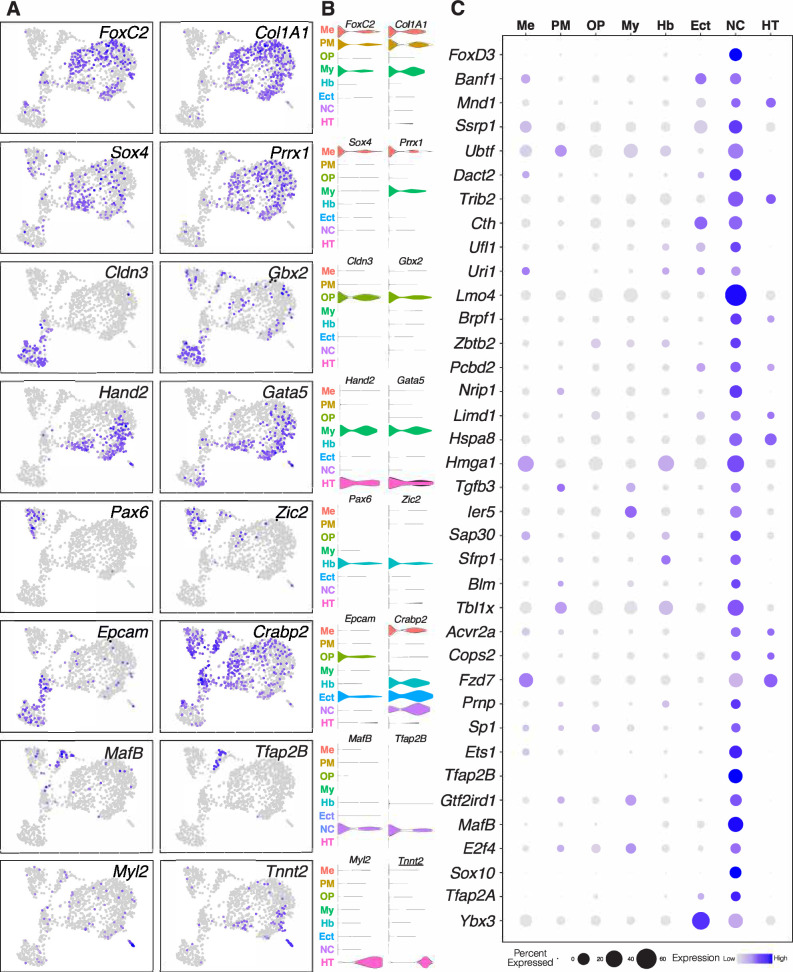
Figure 1. Single-cell (sc) RNA-seq of hindbrain neural crest reveals known and novel transcriptional regulators.
(A) Schematic diagram illustrating the pipeline for performing scRNA-seq on the 10X Genomics platform. Reporter expression mediated by the FoxD3-NC2 enhancer (A’) was used as reference to dissect the hindbrain of HH12 chick embryos. Barcoded GEMs generated from the single-cell suspension were sequenced at a median depth of 50,000 reads/cell. (B) Dimensional reduction using UMAP identifies six subpopulations (including the spike-in) contained within the dissociated embryonic hindbrain. (C) Subset of B showing cells from hindbrain (Hb), ectoderm (Ect), and neural crest (NC). (D–D’) Feature plots used to visualize the expression of known marker genes as a means of identifying subpopulations in (C) in low-dimensional space. Single-cell expression distribution for marker genes (D’) in each cluster is shown as violin plots. (E) Genes that were associated with the GO terms ‘DNA binding’, ‘regulation of transcription,’ or ‘transcription factor’ were characterized as transcriptional regulators and the relative expression and abundance of a subset of them was visualized as a dot plot. The size of each dot corresponds to the percentage of cells expressing that specific gene in a given cluster, while the color represents the average expression level. (F) Feature plots showing expression of previously uncharacterized transcription factors or chromatin remodelers expressed in neural crest cells. (G–K) Hybridization chain reaction was used to validate the expression of a few factors that were identified in (E). Dorsal view of the hindbrain of HH12 shows migratory neural crest streams at r4 and r6 surrounding the otic. Hb, hindbrain; ot, otic placode; r, rhombomere; nc, neural crest; ect, ectoderm. See also Figure 1—figure supplements 1 and 2.