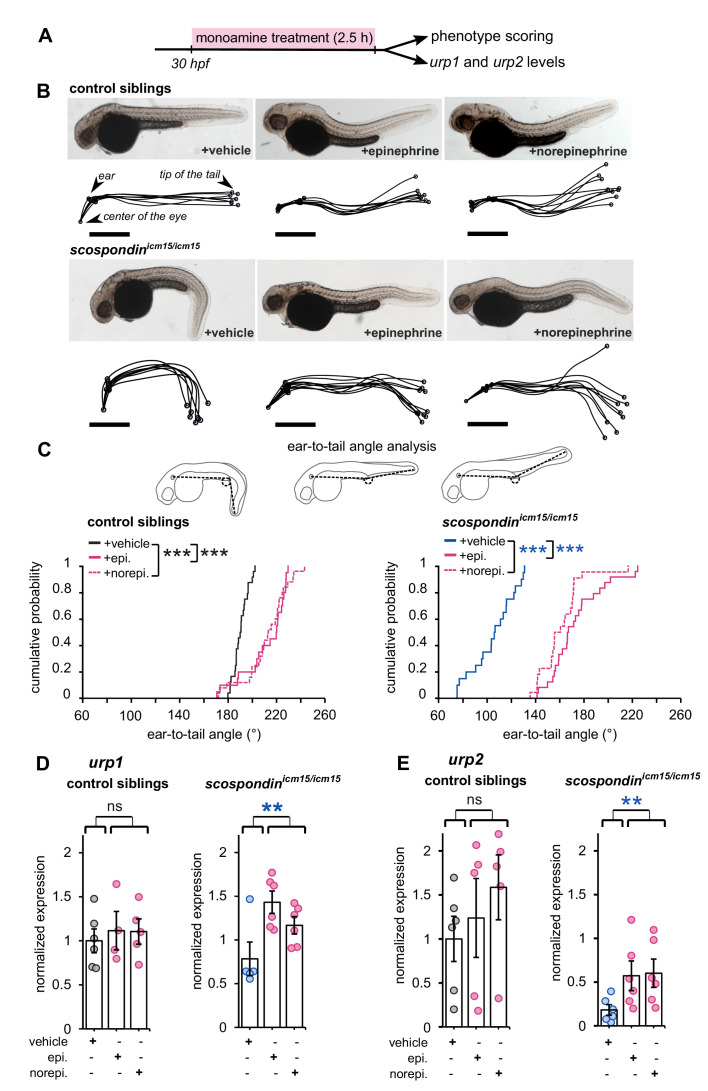
Figure 4. Epinephrine and norepinephrine compensate the loss of the Reissner fiber for body axis straightening and increase urp expression.
(A) Curled-down scospondinicm15/icm15 mutants and their control siblings were sorted at 30 hpf according to the geometry of their posterior axis and then exposed to a E3 solution (vehicle), epinephrine or norepinephrine for 2.5 hr prior to phenotype scoring and RNA extraction. (B) Representative pictures of control siblings (top) and scospondinicm15/icm15 mutants (bottom) after vehicle (left), epinephrine (middle) or norepinephrine (right) treatments. For each condition, the global morphologies of treated embryos are represented by superimposed traces linking the center of the eye, the ear and the tip of the tail in one representative clutch. Scale bar: 0.5 mm. (C) Quantification of the angle formed between the ear, the caudal limit of the yolk extension and the tip of the tail (as shown on the schematics, top) in control siblings (bottom left) and scospondinicm15/icm15 mutants (bottom right). Data were collected from three independent experiments and include 24, 20, 25 control siblings treated with a vehicle solution, epinephrine, norepinephrine respectively (black, solid pink, and dotted pink line respectively) and 20, 24, 22 scospondinicm15/icm15 embryos treated with a vehicle solution, epinephrine, norepinephrine respectively (blue, solid pink, and dotted pink line respectively) ***p<0.001 (Kolmogorov-Smirnov test). (D, E) qRT-PCR analysis of the mRNA level of urp1 (D) and urp2 (E) in control siblings (left) and scospondinicm15/icm15 embryos (right). Data are represented as mean ± SEM. n = 4 to 6 independent replicates for each condition. Each point represents a single experimental replicate. ns p>0.05, **p<0.01 (GLM test).