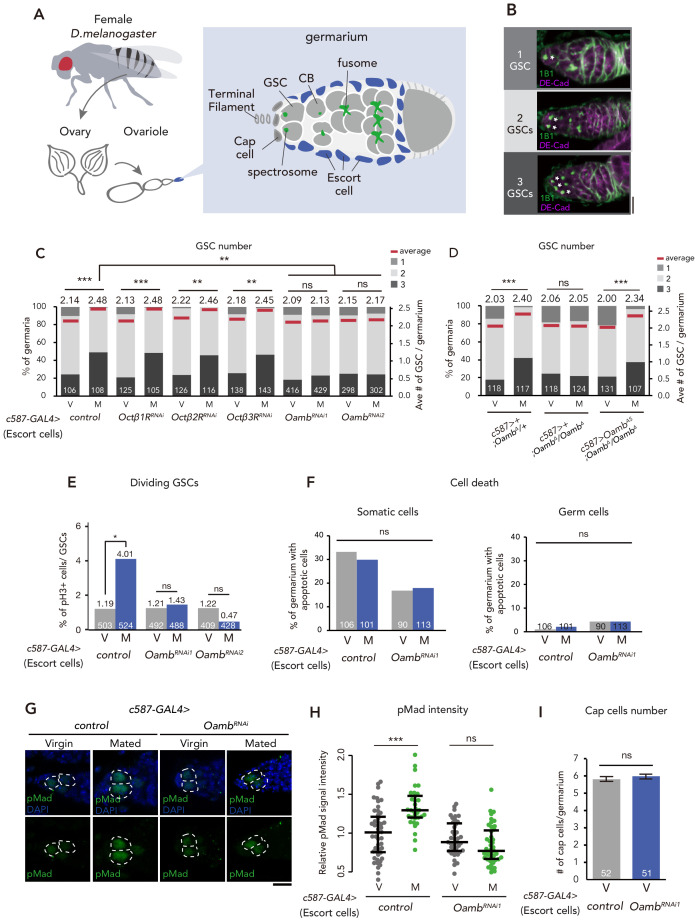
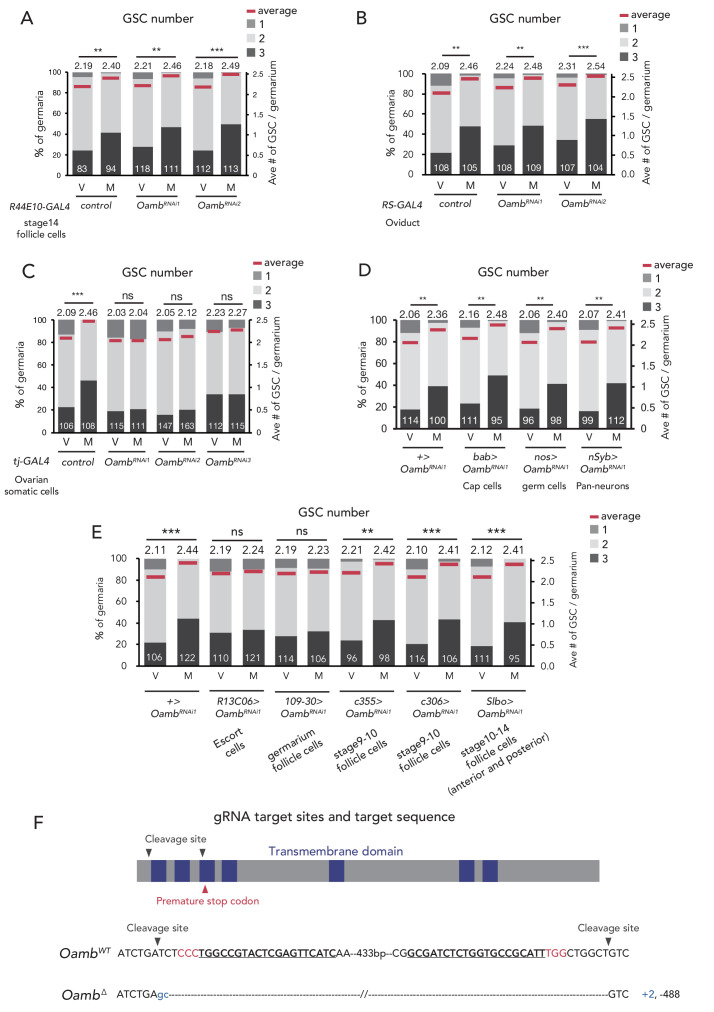
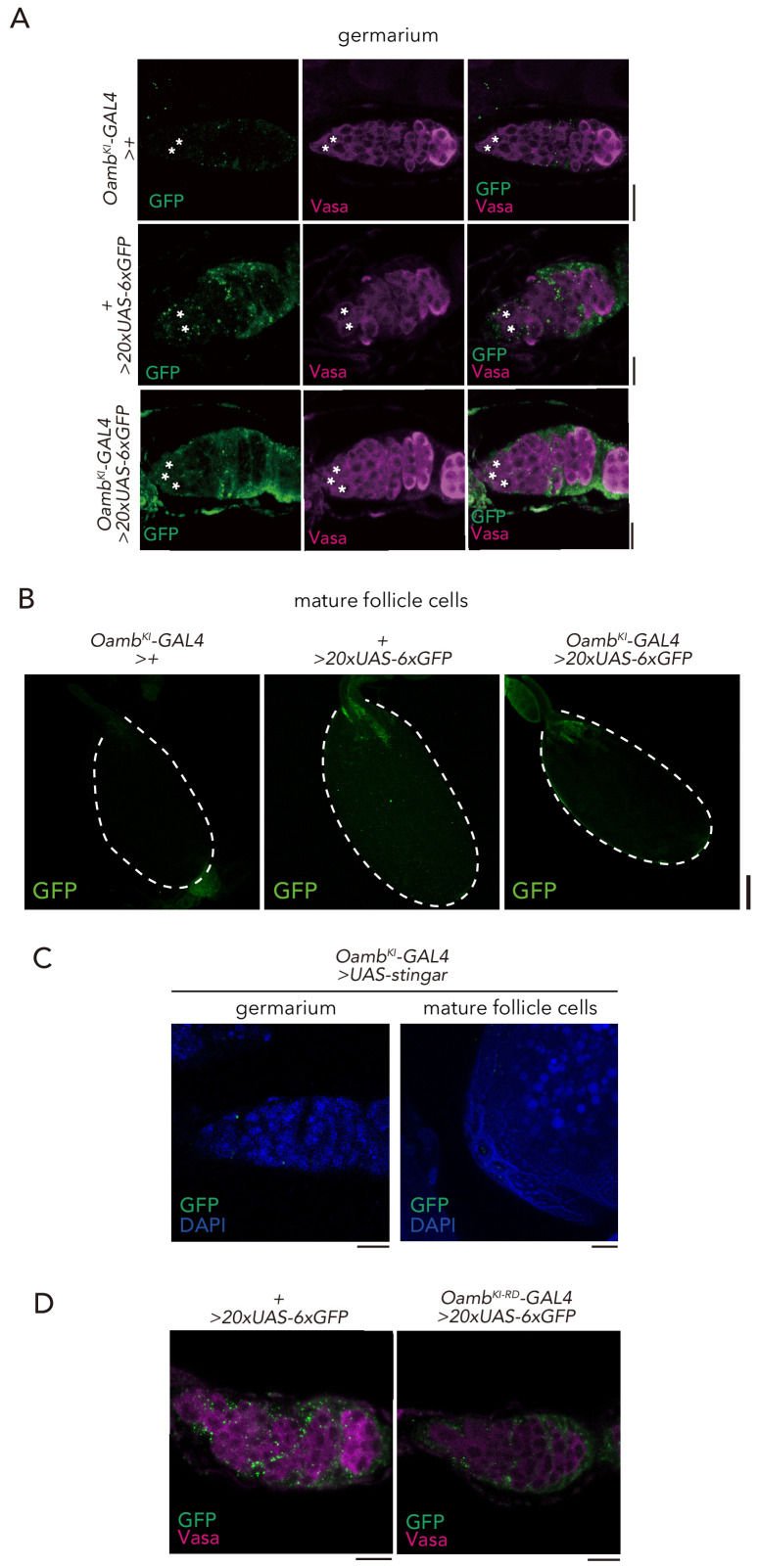
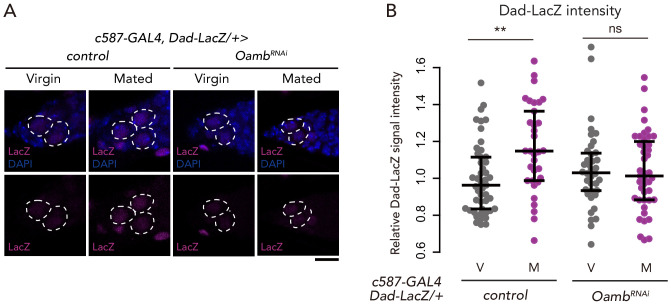
Figure 1. Post-mating GSC increase requires Oamb in the escort cells.
(A) A schematic representation of Drosophila germarium. GSCs reside in a niche consisting of somatic cells such as cap cells, terminal filament cells, and escort cells and are identifiable by their stereotypical spectrosome morphology and location (adjacent to cap cells). GSC division produces one self-renewing daughter and one cystoblast (CB) that differentiates into a germline cyst. (B) Representative images of wild-type (w1118) female adult germariums, containing 1, 2 and 3 GSCs from top to bottom. The samples were stained with monoclonal antibody 1B1 (green) and anti-DE-cadherin (magenta), which stain the spectrosome and overall cell membranes, respectively. GSCs are indicated by asterisk. Scale bar, 20 µm. (C–D) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. c587>+ flies were used as the control in D. (E) The ratio of pH3+ GSCs per total GSCs. (F) The ratio of apoptotic (Dcp-1+) somatic cells and germ cells per germarium. c587>+ flies were used as the control. (G) Representative images of adult female germaria immunostained with anti-pMad antibody (green) and DAPI (blue) are shown. GSCs are outlined with dotted lines. Scale bar, 10 µm. (H) Quantification of relative pMad intensity levels in the GSCs (i.e. virgin (V), mated (M)) as normalized to the pMad intensity in CBs. Each sample number was at least 25. The three horizontal lines for each sample indicate lower, median, and upper quartiles. (I) The number of cap cells per germarium in the control and Oamb RNAi driven by c587-GAL4. Values on the y-axis are presented as the mean with standard error of the mean. c587>+ flies were used as the control. For C-F, and I the number of germaria analyzed is indicated inside the bars. Wilcoxon rank sum test with Holm’s correction was used for C, D, H, and I. Fisher’s exact test with Holm’s correction was used for E and F. ***p≤0.001, **p≤0.01, and *p≤0.05; NS, nonsignificant (p>0.05). All source data are available in Source data 1 and 2.