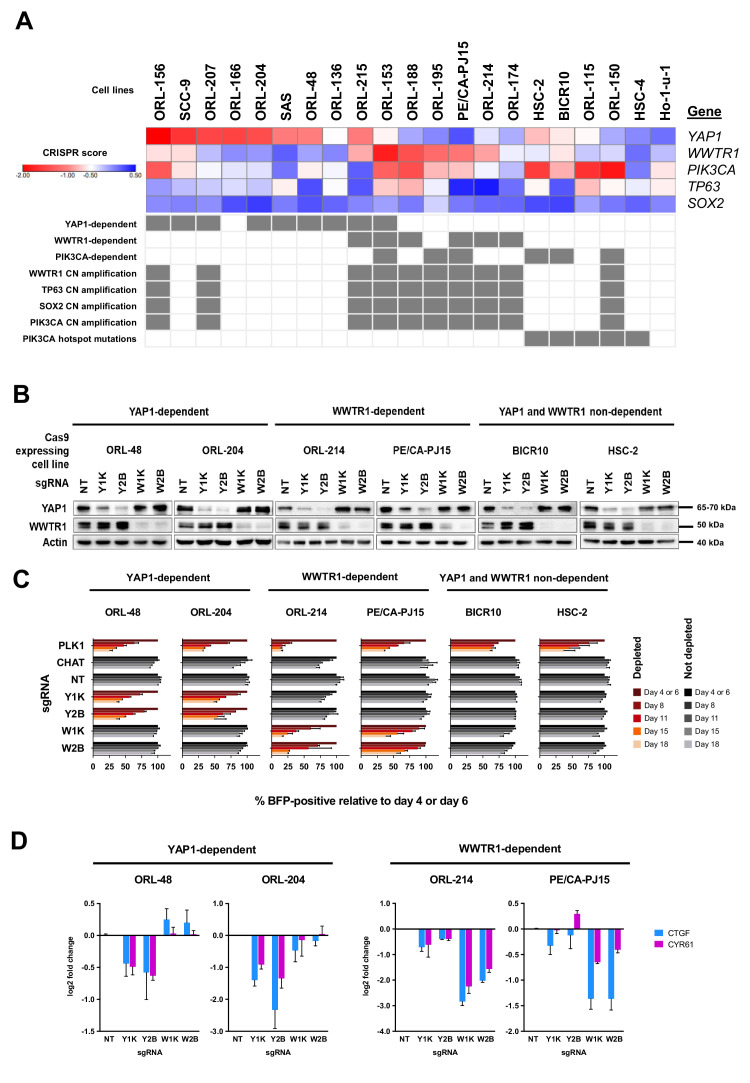
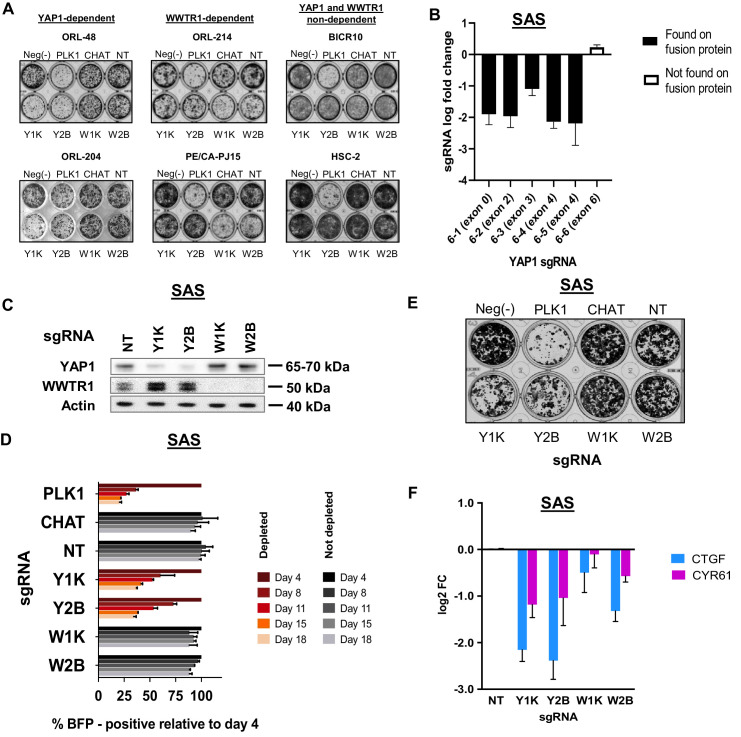
Figure 3. YAP1 and WWTR1 of the Hippo pathway are selectively essential in a distinct subset of OSCC cell lines.
(A) Essentiality profile (depicted with CRISPR scores heatmap) of YAP1, WWTR1, PIK3CA, TP63 and SOX2 across 21 OSCC cell lines derived from the CRISPR/Cas9 screen. Dependency on these genes were depicted as grey box in the bottom panel, according to the MAGeCK definition of significant depletion at FDR ≤ 0.05. No cell lines were dependent on TP63 or SOX2. The degree of essentiality differs across the lines. Some subsets of the cell lines are only dependent on either YAP1 or WWTR1, while neither gene appears to be essential in another subset of cell lines. PIK3CA, TP63 and SOX2 are genes implicated in HNSCC carcinogenesis that are often co-amplified with WWTR1, located on chromosome 3q25-28. All WWTR1-dependent cell lines had copy number amplification on these genes while all PIK3CA mutated cell lines are not dependent on either YAP1 or WWTR1. (B) Western blot images showing the protein level of YAP1 and WWTR1 on day 4 upon transducing the Cas-9 expressing cell lines with lentivirus carrying gene-specific sgRNA. Two sgRNAs were used per target gene. (C) Co-competition assay was used to validate the essentiality of YAP1 and WWTR1 on the selected cell lines. The growth of the BFP-positive transduced population was compared to the non-transduced population throughout the 18 days assay. The percentage of BFP-positive cells obtained at different time points were normalized to the day 4 readings for respective sgRNA (except ORL-204 which had time points normalized to the day 6 readings for respective sgRNA). PLK1 is a core essential gene included as a positive control. Negative controls include CHAT which is a non-essential gene across the panel of cell lines, and NT serves as a non-targeting control. Data are shown as mean ± SD (n = 2 biological repeats). (D) qPCR results show suppression of downstream targets of YAP1 and WWTR1 only when the respective fitness gene is being knocked-out. Down-regulation of CTGF and CYR61 gene expression was observed when YAP1 is knocked-out in the YAP1-dependent cell lines (ORL-48 and ORL-204). In the WWTR1-dependent cell lines (ORL-214, PE/CA-PJ15), CTGF and CYR61 expression is only suppressed when WWTR1 is knocked-out. Data are shown as mean ± SD (n = 2 independent experiments with technical triplicates).