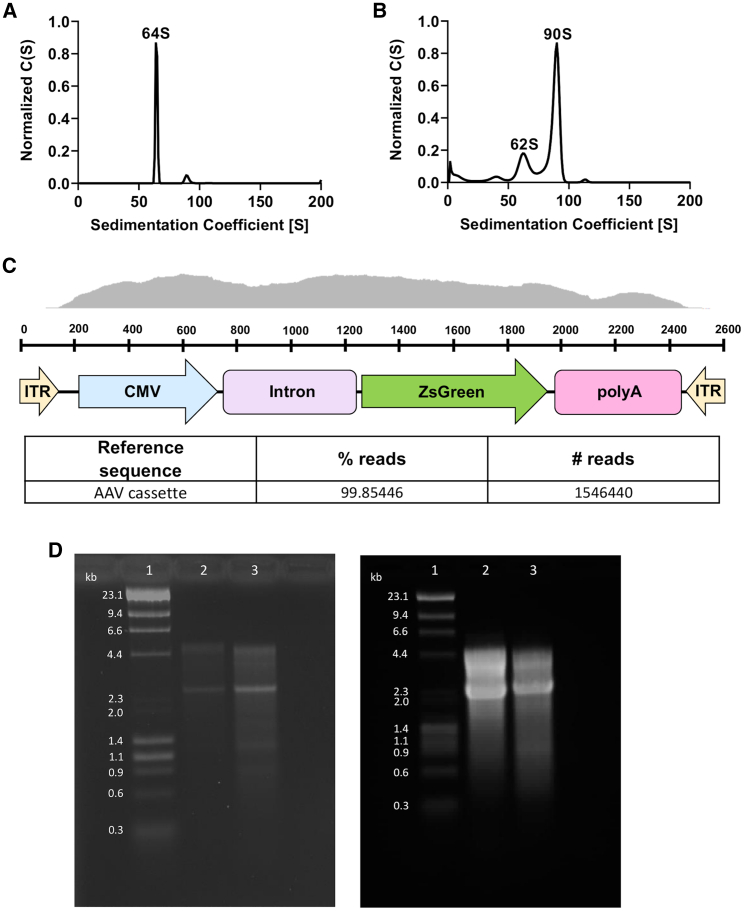
Figure 5.
Analytical Ultracentrifugation-Sedimentation Velocity Profiles of AAV9 Produced in ExpiSf9 Cells and Alkaline Gel Electrophoresis of Insect Cell-Derived AAV9 Genomes
(A) Empty AAV9 capsid was analyzed by AUC in order to identify the lower boundary. (B) Once the lower empty capsid boundary was set, AAV9 containing the transgene cassette was run. Each peak was then integrated using the SEDFIT software to determine the percentage of full viral particles. The sedimentation coefficient on the x axis is in Svedberg units (S). The y axis represents the concentration (C) as a function of S. (C) Diagram showing the sequencing coverage of the recombinant AAV9 genome. Bottom table shows the percentages of the AAV genome in the recombinant AAV9 preparations. (D) Sf9- and ExpiSf9-derived AAV9 ZsGreen genomes were isolated and run on an alkaline gel. Lane 1, DNA ladder; lane 2, Sf9-derived AAV9 ZsGreen genome; lane 3, ExpiSf9-derived AAV9 ZsGreen genome. 500 ng of DNA was loaded on the left gel, and 7,500 ng of DNA was loaded on the right gel. The predicted genome size is ~2.6 kilobase (kb) pairs.