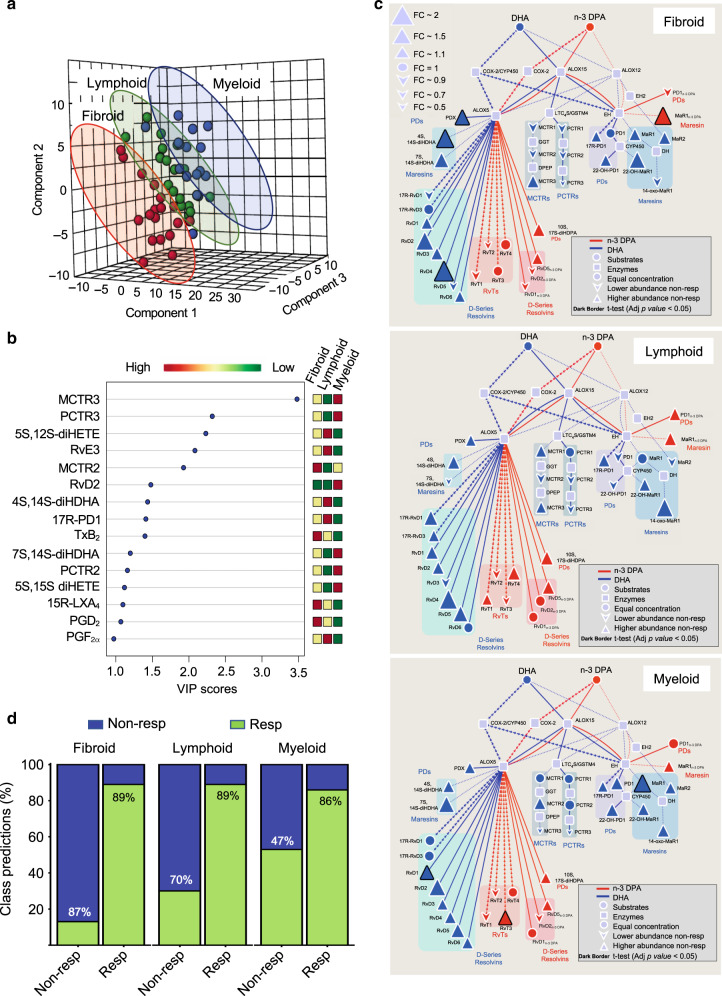
Fig. 3. Combining disease pathotypes and select SPM concentrations enhances model predictiveness.
Plasma was collected from RA patients prior to the initiation of treatment with DMARD and lipid mediator concentrations established using LC-MS/MS-based lipid mediator profiling. a, b PLS-DA analysis of peripheral blood lipid mediator concentrations for lympho-myeloid (lymphoid), diffuse-myeloid (myeloid) and pauci-immune-fibroid (fibroid) pathotypes. a 3-dimensional score plot. b Variable importance in projection (VIP) scores of 15 lipid mediators with the greatest differences in concentrations between the three groups. Results are representative of n = 18 for Fibroid n = 17 for myeloid and n = 19 for lymphoid. c Pathway analysis for the differentially expressed mediators from the DHA and n-3 DPA bioactive metabolomes in DMARD non-responders (Non-Resp) when compared to DMARD responders (Resp) for each pathotype. Statistical differences between the normalised concentrations (expressed as the fold change) of the lipid mediators from the Non-Resp and Resp groups were determined using two-sided t test followed by a multiple comparison correction using Benjamini–Hochberg procedure. Up- or downregulated mediators are denoted with using upward and downward facing triangles, respectively, and on changes of the node’s size. Bolded mediators represent statistical differences between the two groups when adjusted p value <0.05. Results are representative of n = 18 for fibroid Resp, n = 15 for fibroid Non-Resp, n = 19 for lymphoid Resp, n = 10 for lymphoid Non-Resp, n = 22 for myeloid Resp, n = 15 for myeloid Non-Resp. d Classification accuracies for each class (sensitivity and specificity) of the RvD4, 10S,17S-diHDPA, 15R-LXA4 and MaR1n-3 DPA model created using the specific dataset for the different pathotypes (fibroid, lymphoid and myeloid). Green indicates the samples that were predicted as Resp while blue indicates predicted Non-Resp. Percentages indicate true positives (Resp class) and true negatives (Non-Resp class). All the models were created using the random forest methodology (“randomForest” package from R). Source data are provided as a Source data file.