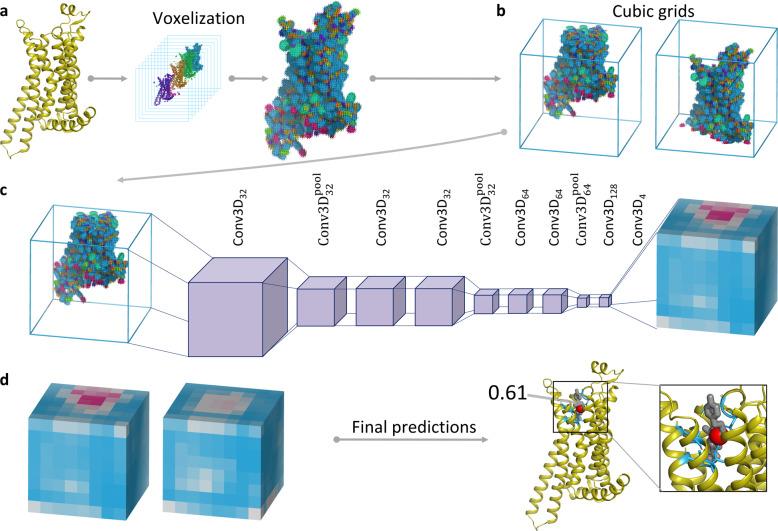
Fig. 1. Schematic representation of the BiteNet workflow.
a The input three-dimensional structure of a protein is represented with voxel grid, where channels correspond to the atomic densities. b The voxel grid is split into fixed-size cubic grids to be fed a neural network. c Each cubic grid is processed with the 3D convolutional neural network to predict binding sites in fixed-size cells. Cells in cubic grids are colored according to the probability score confidence, from blue to red. d Predictions obtained for each cubic grid are then processed to output center of the binding site (red sphere), its probability score, and amino acid residues within 6 Å of the predicted center (blue sticks). Co-crystallized ligand is shown with gray sticks.