Abstract

Proteins often have both orthosteric and allosteric binding sites. Endogenous ligands, such as hormones and neurotransmitters, bind to the orthosteric site, while synthetic ligands may bind to orthosteric or allosteric sites, which has become a focal point in drug discovery. Usually, such allosteric modulators bind to a protein noncompetitively with its endogenous ligand or substrate. The growing interest in allosteric modulators has resulted in a substantial increase of these entities and their features such as binding data in chemical libraries and databases. Although this data surge fuels research focused on allosteric modulators, binding data is unfortunately not always clearly indicated as being allosteric or orthosteric. Therefore, allosteric binding data is difficult to retrieve from databases that contain a mixture of allosteric and orthosteric compounds. This decreases model performance when statistical methods, such as machine learning models, are applied. In previous work we generated an allosteric data subset of ChEMBL release 14. In the current study an improved text mining approach is used to retrieve the allosteric and orthosteric binding types from the literature in ChEMBL release 22. Moreover, convolutional deep neural networks were constructed to predict the binding types of compounds for class A G protein-coupled receptors (GPCRs). Temporal split validation showed the model predictiveness with Matthews correlation coefficient (MCC) = 0.54, sensitivity allosteric = 0.54, and sensitivity orthosteric = 0.94. Finally, this study shows that the inclusion of accurate binding types increases binding predictions by including them as descriptor (MCC = 0.27 improved to MCC = 0.34; validated for class A GPCRs, trained on all GPCRs). Although the focus of this study is mainly on class A GPCRs, binding types for all protein classes in ChEMBL were obtained and explored. The data set is included as a supplement to this study, allowing the reader to select the compounds and binding types of interest.
Introduction
Drugs bind to proteins to exert a biological effect, which can be activation, deactivation, or blockage to prevent an endogenous ligand from binding. The location on the protein where the endogenous, or natural, ligand binds is called the orthosteric binding site. All other binding sites are described as allosteric binding sites.1 Allosteric binding of ligands (allosteric modulation) has been reported and studied at many protein classes, including membrane-bound G protein-coupled receptors (GPCRs).1−3 Moreover, structural information is available from the Protein DataBank,4 which shows the binding of allosteric ligands to GPCRs at different sites (Figure 1).2,5 The transmembrane (TM) domains of the orthosteric binding pocket in GPCRs are highly conserved for most GPCR classes. An exception is formed by class C GPCRs, for which the orthosteric site lies in the extracellular Venus flytrap domain. The most common allosteric binding site for class C GPCRs is located close to or at the orthosteric site in the TM domain of other (class A) GPCRs.6 Other GPCR allosteric binding sites have been identified close to or in the intracellular domain and between helices in the TM domain.2,3 The physicochemical properties of these multiple binding sites can vary greatly, enabling dissimilar ligands to bind the same protein via different sites.
Figure 1.
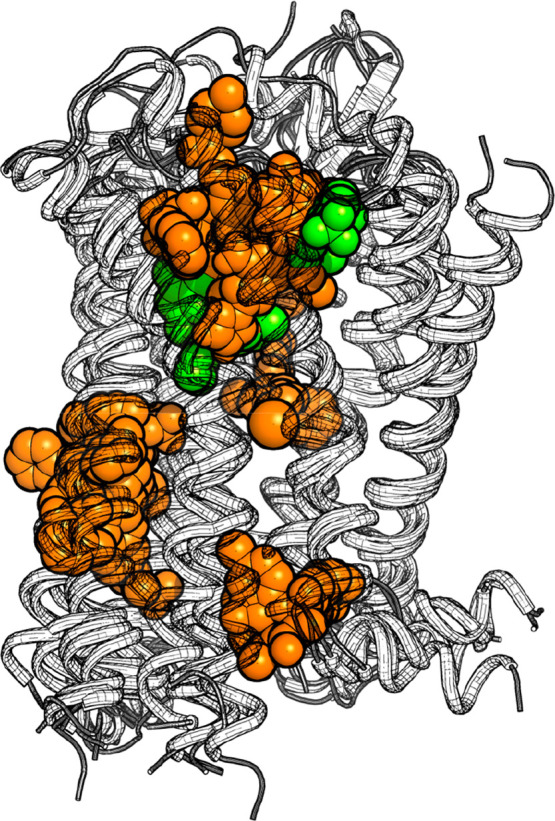
Ligand binding sites observed in crystal structures of class A GPCRs. Orthosteric ligands are shown in green, allosteric modulators in orange. The following crystal structures are included (PDB): 4MBS,10 4MQT,10 4N6H,11 4NTJ,12 4PHU,13 5LWE,14 5NDZ,15 5NLX,16 5T1A,2 5TZR,17 5TZY,17 5X7D,18 6C1Q,19 and 6C1R.20
Although allosteric modulators are widely studied and reported, allosterism of compounds is not yet annotated in the public chemical database ChEMBL.7 Previous attempts have been made to retrieve allosteric properties of compounds from the original published article to fill this information gap in the ChEMBL database.8,9 However, although allosteric and orthosteric annotations were retrieved, the added value of this information in binding predictions was not assessed. We observed that allosteric compounds bind with a lower absolute affinity on average but with similar ligand efficiency, and we hypothesized that the inclusion of this information would lead to a better prediction of affinity.8
In the current study an updated text mining protocol is presented that annotates orthosteric and allosteric compounds more accurately. Moreover, the data set is additionally curated for class A GPCRs, improving the data set’s quality for this target type. The value of the allosteric annotation of compounds was assessed by training pChEMBL prediction models for class A GPCRs with and without binding type information. Furthermore, models were constructed that were able to predict allosterism of class A GPCR compounds. The entire annotated data set and the script for model training are freely available as a supplement to this article. Additionally, the reader is able to apply text mining on new documents to annotate the binding type of compounds by using the text mining method and keywords that are provided.
Results
Construction of the Orthosteric/Allosteric Binding Type Data Set Using Text Mining
Keyword-enabled text mining was applied on documents retrieved from ChEMBL7 to determine the binding type of the compounds (see Methods for details). Only the compounds from the assays that were annotated as “primary assay” were categorized as allosteric, orthosteric, bitopic, or covalent. The primary assay was assigned based on the highest number of compounds per assay of each document. In contrast to previous work where an effort was made to characterize allosteric compounds,8,9 the bitopic and covalent classes were added to remove these compounds from the allosteric and orthosteric classes. Upon manual assessment of the text-mined orthosteric and allosteric compounds for class A GPCRs, it was observed that one document contained a primary assay with computationally derived activity values instead of experimental binding activities.21 Therefore, compounds from the secondary assay were selected for this document. Furthermore, 46 histamine H4 compounds that were categorized as allosteric were in fact orthosteric, which was detected when checked manually. These compounds were corrected accordingly. To expand the data set, additional “undetermined” class A GPCR documents were manually categorized, which resulted in a further addition of 306 orthosteric and 24 allosteric compounds. For the majority the binding types were well-defined, especially for class A GPCRs, as confirmed by manual random sample assessment and positive model outcomes. The total binding type data set (for all text-mined proteins) contained 201 721 data points (9 390 allosteric, 14 265 orthosteric, and 178 066 undetermined), of which 60 116 defined data points for class A GPCRs (1 442 allosteric, 9 914 orthosteric, and 48 760 undetermined).
Chemical Properties of Allosteric Modulators
The differences between orthosteric and allosteric binders were explored by evaluating the following properties: molecular weight, A Log P, number of hydrogen bond donors and acceptors, activity (pChEMBL), number of rotatable bonds, and number of aromatic bonds. It was observed that the properties of the allosteric modulators for all proteins in ChEMBL were comparable to those of the orthosteric compounds (Table 1). In general, the orthosteric compounds covered a wider range of all properties: additional higher molecular weights, more diversity in A Log P, and compounds with more hydrogen bond donors and acceptors. A slight difference was observed in the spread of bioactivities of the two binding types. Although the orthosteric compounds included a higher number of low pChEMBL values (pChEMBL < 4), the bioactivity median was 0.7 log units higher than that of allosteric compounds (pChEMBL 7.1 and 6.4, respectively), in line with previous work.8 Moreover it was observed that allosteric modulators contained a bigger fraction of aromatic bonds (value normalized to the number of bonds) compared to orthosteric compounds. This corresponds with previous statements that allosteric modulators are more aromatic.8,9 Please note that for the modeling work below, a more diverse number of descriptors was used. Nonetheless, the physicochemical properties of the allosteric modulators are generally not very different from those of orthosteric compounds.
Table 1. Physicochemical Properties of Orthosteric and Allosteric Compounds in ChEMBLa.
| allosteric |
orthosteric |
|||||
|---|---|---|---|---|---|---|
| median | MAD | mean and standard deviation | median | MAD | mean and standard deviation | |
| pChEMBL | 6.4 | 0.9 | 6.5 ± 1.2 | 7.1 | 1.0 | 7.1 ± 1.4 |
| molecular weight | 383 | 61 | 404 ± 135 | 434 | 93 | 541 ± 474 |
| A Log P | 3.8 | 1.0 | 3.8 ± 2.0 | 3.8 | 1.3 | 3.1 ± 3.9 |
| num. H donors | 1 | 1 | 1 ± 2 | 1 | 1 | 4 ± 8 |
| num. H acceptors | 4 | 1 | 4 ± 2 | 5 | 2 | 6 ± 8 |
| fraction rotatable bonds | 0.17 | 0.05 | 0.18 ± 0.09 | 0.20 | 0.06 | 0.22 ± 0.12 |
| fraction aromatic bonds | 0.50 | 0.11 | 0.47 ± 0.17 | 0.43 | 0.12 | 0.42 ± 0.18 |
The fractions of rotatable bonds and aromatic bonds represent the number of bonds normalized to the total number of bonds per compound. An independent (unpaired) t test gave p < 0.001 (two-tailed, α = 0.05) for the difference between the allosteric and orthosteric means of each property, which indicates a significant difference. MAD = mean absolute deviation.
When only a specific protein class was explored, class A GPCRs in this case, it was observed that the properties of the orthosteric and allosteric compounds were comparable to the mean of compounds for all proteins in ChEMBL. However, we noticed that the properties of individual protein classes could vary significantly compared to the properties of the entire set. Figure 2 (and Supporting Information Figure S1) displays the variances between the ligand properties of different protein classes. Although class A GPCRs contained the second-largest number of mined allosteric modulators (1442 compounds) and are therefore able to cover more chemical ground, it is remarkable that most protein classes have less pronounced properties: the minimum and maximum values for especially molecular weight, A Log P, and hydrogen bond donors and acceptors were less prominent than for the class A GPCRs. The A Log P values of the orthosteric class B and C GPCR compounds (A Log P 5.2 and A Log P −2.7, respectively) were deviating a lot from the conclusion drawn from the entire set where the median A Log P was 3.8. However, these observations could be explained, as shown below.
Figure 2.
Physicochemical properties of orthosteric and allosteric compounds in ChEMBL per protein family. Orange boxes indicate the area between the 25th percentile and 50th percentile, gray indicates the area between the 50th percentile and 75th percentile, the border between orange and gray indicates the median value or 50th percentile, and the whiskers indicate the minimum value up to the 25th percentile (bottom whiskers) and the 75th percentile up to the maximum value (top whiskers). (A) pChEMBL value; (B) A Log P; (C) fraction of rotatable bonds; (D) fraction of aromatic bonds.
It was observed that the biggest allosteric set was retrieved for class C GPCRs (2513 compounds), of which the orthosteric site is located in the extracellular Venus flytrap domain, in contrast to other GPCR classes. The orthosteric compounds in this class had a observably lower A Log P than the allosteric modulators: A Log P median orthosteric −2.7 and allosteric 3.7. Given that the orthosteric compounds for class C GPCRs bind to a structurally different binding site than the allosteric modulators for this receptor, this observation is not surprising. Furthermore, it is known that glutamate receptors make up a large fraction of class C GPCRs, for which the main natural ligand is glutamate, a charged amino acid.22 Hence, orthosteric ligands should resemble these characteristics, leading to the observed shift in A Log P values (the median A Log P of orthosteric compounds binding to glutamate receptors is −3.0). Moreover, it is noteworthy to mention that the class C GPCR compounds might bias the full data set as they represent 27% of the allosteric modulators. A similar trend was observed for class B GPCRs, with an A Log P median for orthosteric compounds of 5.2. It was found that the orthosteric compounds for class B GPCRs consisted of solely peptides and that the allosteric class was only represented by seven modulators, which were also peptides. Therefore, class B GPCRs alone will not be a good case for modeling allosterism.
Class A GPCRs, on the other hand, contained a sufficient number of orthosteric and allosteric compounds (9914 orthosteric and 1442 allosteric) and a well-balanced portion of small molecules and peptides: orthosteric 5428 small molecules, 4395 peptides, and 91 undefined; and allosteric 851 small molecules and 591 peptides (as classified by ChEMBL).23 It should be noted that the results in this section are based on text-mined binding types. Therefore, caution is advised when comparing and drawing conclusions from the differences observed between orthosteric and allosteric compounds and also between protein classes. Nonetheless, an effort was made to curate the binding types for class A GPCRs with higher accuracy to support the findings in the current study.
Chemical Structures of Class A GPCR Allosteric Modulators
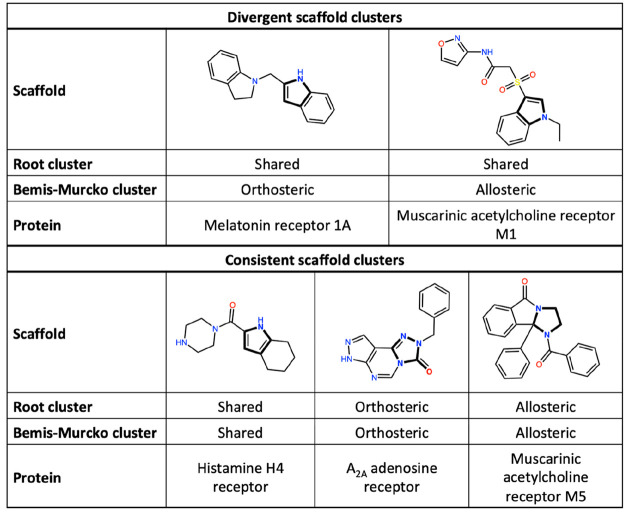
The chemical structures of compounds for the class A GPCRs were explored based on their scaffold trees. The scaffolds were clustered based on two tree levels: the root (smallest scaffold) and the Bemis–Murcko framework24 (biggest scaffold). Clustering of the class A GPCR set resulted in 313 unique root scaffolds and 4362 Bemis–Murcko scaffolds. The allosteric modulators covered 19 root scaffolds and 564 Bemis–Murcko scaffolds. The orthosteric compounds contained significantly more unique scaffolds: 229 root and 3770 Bemis–Murcko scaffolds. Furthermore, it was observed that some clusters contained both orthosteric and allosteric compounds: 65 root and 28 Bemis–Murcko scaffold clusters were shared. Notably, clusters formed based on the root scaffold are not uniform with the clusters based on Bemis–Murcko scaffolds. In contrast, it was observed that compounds that were grouped into a shared cluster based on root scaffold were clustered into either an orthosteric-specific or an allosteric-specific cluster based on the Bemis–Murcko scaffold (Figure 3). This can be explained since the Bemis–Murcko scaffolds are bigger and therefore more specific. The root scaffold, however, is more general and therefore prone to cluster more diverse compounds into the same cluster.
Figure 3.
Clustered scaffolds of orthosteric and allosteric compounds for class A GPCRs. Bemis–Murcko scaffolds are depicted with the root scaffold indicated in bold.
Predicting Allosteric Modulation
Convolutional deep neural networks (DNNs, see Methods for network architecture) were applied to observe if allosteric modulation of compounds for class A GPCRs could be predicted. Compounds of the binding type data set that were published before the year 2013 were used for model training. Compounds published in 2013 or later were used for validation. This time-split, or temporal-split, approach has advantages over random-split validation as it gives a more accurate representation of the models’ predictive performances.25 Furthermore, the training set was balanced to correct for the unequal number of orthosteric and allosteric compounds. This was performed using oversampling of the minority class. The minority class was multiplied until it reached an amount of compounds that was comparable to the majority class.
The allosteric and orthosteric binding types were predicted by a model trained on physicochemical properties, compound fingerprints, and protein descriptors (for details see Methods). The temporal-split validation showed that the proteochemometric (PCM)26 DNN model, which was trained on compound and protein class A GPCR data, could be used to differentiate allosteric modulators from orthosteric compounds: Matthews correlation coefficient (MCC) = 0.54, sensitivity = 0.54, specificity = 0.94, positive predictive value (PPV) = 0.82, and negative predictive value (NPV) = 0.80. Although sensitivity for allosteric compounds is significantly lower than that for orthosteric compounds (the specificity metric in this case), the model’s predictive ability is sufficient to differentiate between both binding types.
DNNs Outperform Random Forest in Binding Type Predictions
The binding type DNN model was compared to a random forest (RF) model that was trained and tested on exactly the same data (class A GPCRs). The predictive performance of the RF model for allosteric compounds was slightly worse than that of the DNN model with a performance of MCC = 0.43, sensitivity = 0.35, specificity = 0.97, PPV = 0.85, and NPV = 0.74. This result indicates that the DNN model was better than the RF model in identification of the binding type of class A GPCRs.
Enhancement of Bioactivity Predictions by the Incorporation of Binding Types
It was studied if the text mining derived binding types could contribute to the predictive performance of pChEMBL prediction models. Instead of the prediction of binding types, the DNN models were trained to predict the activity of compounds (pChEMBL value ≥6.5 is active), using the binding types as descriptor. The models were validated using the same temporal split as mentioned previously (split on the year 2013). The effect of the implementation of binding types was analyzed by comparison of the performances of the pChEMBL prediction model with and without binding type. The models were trained on both orthosteric and allosteric compounds but validated on allosteric modulators and orthosteric compounds separately to remove any bias that might result from the addition of the binding type descriptors. Potential bias might be induced since the allosteric compounds are on average less potent than the orthosteric compounds: pChEMBL 6.6 ± 1.3 and 7.3 ± 1.3, respectively, for class A GPCRs. The findings for bioactivity predictions for allosteric modulators are reported in this section and Table 2, and the results for orthosteric compounds are included in Supporting Information File S2. It was observed that the MCC of the class A GPCR pChEMBL prediction model with implemented binding type descriptors was slightly higher than for the pChEMBL prediction model that lacked these descriptors, MCC 0.27 and 0.30, respectively, for allosteric modulators. Remarkably the overall performance (MCC) increased slightly for allosteric bioactivity predictions, but sensitivity decreased (−0.05) when adding binding types. Nevertheless, specificity and accuracy both increased: specificity +0.12 and accuracy +0.03. The receiver operating characteristic (ROC) remained comparable (or slightly worse −0.01).
Table 2. DNN Regression Model Performances for Prediction of Bioactivities of Allosteric Modulators for Class A GPCRsa.
| training data set | added descriptor | MCC | sensitivity | specificity | accuracy | PPV | NPV | RMSE | ROC |
|---|---|---|---|---|---|---|---|---|---|
| class A GPCRs | – | 0.27 | 0.81 | 0.44 | 0.61 | 0.55 | 0.74 | 1.07 | 0.73 |
| binding type | 0.30 | 0.74 | 0.56 | 0.64 | 0.58 | 0.72 | 1.15 | 0.72 | |
| predicted binding type | 0.22 | 0.51 | 0.70 | 0.62 | 0.59 | 0.63 | 1.01 | 0.70 | |
| GPCRs | – | 0.27 | 0.78 | 0.48 | 0.62 | 0.56 | 0.73 | 1.01 | 0.74 |
| binding type | 0.34 | 0.78 | 0.55 | 0.65 | 0.59 | 0.75 | 1.09 | 0.73 | |
| predicted binding type | 0.15 | 0.52 | 0.63 | 0.58 | 0.54 | 0.61 | 0.99 | 0.67 | |
| all proteins in ChEMBL | – | 0.35 | 0.74 | 0.61 | 0.67 | 0.61 | 0.74 | 1.03 | 0.74 |
| binding type | 0.37 | 0.74 | 0.62 | 0.68 | 0.62 | 0.74 | 0.98 | 0.75 | |
| predicted binding type | 0.18 | 0.43 | 0.74 | 0.60 | 0.58 | 0.61 | 1.06 | 0.67 |
MCC = Matthews correlation coefficient, PPV = positive predictive value, NPV = negative predictive value, RMSE = root-mean-square error, and ROC = receiver operating characteristic.
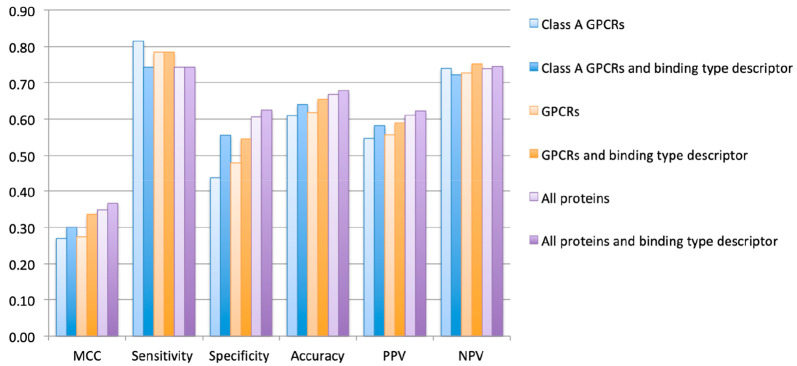
The addition of the binding types and their effect on pChEMBL predictions for allosteric modulators were further evaluated when the model was trained on the full GPCR data set and on the data set containing all proteins in ChEMBL. The pChEMBL predictions for class A GPCRs increased when binding types were included in the models trained on these bigger data sets (Figure 4). All metrics improved or performed equally upon the addition of binding types to these enlarged data sets. The pChEMBL predictions for allosteric modulators of class A GPCRs when trained on the full GPCR data set improved the most with a MCC of 0.27 increasing to 0.34. The best performing model for allosteric modulators for class A GPCRs was the PCM model trained on all proteins with binding types included (MCC 0.37).
Figure 4.
Performances of DNN regression models for the prediction of bioactivities of class A GPCR allosteric modulators with and without binding type descriptors. MCC = Matthews correlation coefficient, PPV = positive predictive value, and NPV = negative predictive value.
Expansion of Binding Type Predictions into Bioactivity Models
It was observed that the DNN models were able to predict if compounds were binding in an allosteric or orthosteric manner. Furthermore, the addition of binding type descriptors into pChEMBL prediction models enhanced model performance. Therefore, models were applied iteratively by first predicting the binding types of compounds, subsequently followed by implementation of these predictions into training of a pChEMBL prediction model. The compounds used for testing were retrieved from ChEMBL assays that were categorized as “undetermined” by the text mining protocol. The predicted binding types were added as a descriptor which ranged from zero (“0”) to one (“1”), with 1 being allosteric and 0 being orthosteric. The scores for this descriptor were derived as output from the binding type classification model and indicated the probability of a compound belonging to the orthosteric class (score <0.5) and allosteric class (score >0.5). The trained class A GPCR pChEMBL prediction model (regression) was compared to a pChEMBL prediction model that lacked these predicted binding type descriptors. The performance was MCC 0.22, sensitivity 0.51, specificity 0.70, accuracy 0.62, PPV 0.59, NPV 0.63, root-mean-square error 1.01 (RMSE), and ROC 0.70 (Table 2). The pChEMBL prediction model that included predicted binding types overall performed worse than the pChEMBL prediction model without the predicted binding types, with the exception of increased specificity and PPV. This may be a result of the implementation of “uncertain” values to generate new predictions; an additional modeling error is introduced into the pChEMBL prediction model that uses predicted descriptors. Furthermore, it was observed that the performances of the models that included predicted binding types decreased when the training set was enlarged with all GPCRs and all proteins in ChEMBL. This is in contrast with the previous observation that model performance increased upon training set enlargement. This indicates that the binding type descriptor weighs heavily in the determination of the pChEMBL value. Therefore, it is of importance that accurate experimentally validated binding types are used in model training, as predicted binding types are not sufficient.
Discussion
Via our text mining protocol we were able to retrieve accurate “orthosteric” and “allosteric” binding types. The addition of these literature mined binding types in pChEMBL prediction models increased model performance. However, an important aspect is that the binding types should be reliable. Implementation of predicted binding types as a descriptor, with a potential error in prediction, did not improve model performance. Although the binding type model did not enhance the accuracy of pChEMBL predictions, the model could be applied to indicate possible allosterism of compounds, as the performance of the binding type model (MCC = 0.54) is remarkably better than random (MCC = 0). In contrast to our study, allosterism predicting models that are reported in the literature are not specified to class A GPCRs or report a less reliable validation method (random split instead of temporal split).8,27 Therefore, the performance of these models cannot be compared directly.
The physicochemical properties of allosteric modulators generally match the properties of orthosteric compounds; both are druglike.8 Nevertheless, the chemical features can be distinctive between protein classes, which is reflected by the differences in A Log P, fraction of rotatable bonds, and fraction of aromatic bonds.9 For class A GPCRs, root and Bemis–Murcko scaffolds were identified that specifically match allosteric modulators or orthosteric compounds. Additionally, scaffolds were found that were shared among allosteric and orthosteric compounds.
Manual evaluation of the text mining results showed that some documents were misclassified. This indicates that the text mining derived binding types are not 100% accurate and therefore might still contain some inaccuracies. However, the majority of the binding types are well-defined, especially for class A GPCRs, as confirmed by manual random sample assessment and positive model outcomes. Furthermore, the addition of specific keywords to filter for covalent and bitopic compounds increases the accuracy of categorization of the allosteric and orthosteric classes. Although addition of a bitopic class in text mining for allosteric modulators has been reported before,8 implementation of a filter for covalent compounds is only presented in this study. The effort that was made to text mine for binding types has the advantage that multiple binding types can be assembled and annotated. In contrast to the compounds published in the Allosteric Database,28 which only lists allosteric molecules, orthosteric compounds were also actively annotated in this study and thus available as an additional subset.
The data set that resulted from text mining was not limited to class A GPCRs but contained determined binding types for multiple protein classes in ChEMBL including receptors, enzymes, and solute carriers. Furthermore, some compounds could not be divided into one of the assigned binding type classes, resulting in “undetermined binding type” compounds. These compounds were thus not picked up using the extensive list of keywords. The expansion of the searched text from title and abstract text to full paper text may decrease the number of undetermined compounds. However, feasibility may be difficult as not all papers are open access.
It was shown that the expansion of the training set using all proteins increased pChEMBL predictions for class A GPCRs, a finding that has been observed for PCM modeling previously.26 Moreover, implementation of the text mining derived binding types increased model performance even further. Previous research shows that the choice of descriptors can influence model performance.29 Nevertheless, to the best of the authors’ knowledge, the use of binding types as additional descriptors in bioactivity modeling has not yet been reported.
Conclusions
We successfully retrieved allosteric and orthosteric binding types using text mining. Implementation of these binding types into class A GPCR pChEMBL prediction models improved model performance. Although we were able to predict the binding type of compounds, implementation of these predicted binding types into pChEMBL prediction models did not enhance model performance. Since we did observe improvement of model performance when accurate binding types were included, we encourage the use of binding type descriptors in computational models. Unfortunately, binding type information on compounds is not always readily accessible, especially in the public domain. This research therefore stresses the importance of binding type integration in public databases.
Methods
Text Mining and Compound Classification
Binding types, orthosteric or allosteric, were assigned to the compounds using text mining. Documents that contained both a title and an abstract were retrieved from the ChEMBL database (version 22).23,30,31 The abstracts and titles were screened for keywords to classify the corresponding document in one of the following classes: orthosteric, allosteric, bitopic, and covalent. Documents were classified using a three-layer approach. Remaining, or unclassified, documents were termed “undetermined”. Every layer of the classification process included a different set of keywords (Supporting Information datasheet S3), with the most accurate and general keywords in the first layer and decreased accuracy of keywords in the following layer. The third layer contained general orthosteric keywords that were not applied in the first layer as they were generally only mentioned in conjunction with allosteric keywords. The documents that could not be classified within the first layer continued to the second layer and, if applicable, to the third layer. Every document that contained at least one bitopic or covalent keyword was directly classified into the corresponding class. Classification of orthosteric and allosteric documents was determined by majority vote based on the number of occurrences of keywords belonging to the orthosteric or allosteric class. Compounds from the primary assay, the assay containing the most bioactivities, of the orthosteric and allosteric classified documents were retrieved from ChEMBL and assigned to the same class as the corresponding document. Additionally, class A GPCR compounds from the primary assays of previously undetermined documents were placed in the orthosteric class if published before 2000, as from this year on the literature started more actively reporting allosteric binding types. Therefore, we assumed that the study was performed for orthosteric binders, when allosterism was not explicitly mentioned in a paper in the years prior to 2000. The text mining protocol to categorize documents is included as Supporting Information file S4.
Data Set
The ChEMBL7 data derived through text mining were filtered on confidence score 5 (multiple direct proteins), 7 (direct protein complex subunits), and 9 (direct single protein). Additionally, values retrieved from PubChem32 and compounds without a defined or exact pChEMBL value (relation is “=”) were discarded. Only pChEMBL values that were determined using binding assays were kept. Mean pChEMBL values were calculated for bioactivities of duplicate compounds tested on the same target. All compounds were standardized using Pipeline Pilot (version 16.2.0.58):33 the largest fragment was kept, and stereochemistry and charges were standardized. The data set is included as Supporting Information file S5.
Compound Descriptors
Compounds were encoded by their physicochemical properties (Supporting Information file S6). Additionally FCFP6 fingerprints34 were used to describe the compounds’ structures. Moreover, the activity types (Ki, Kd, EC50, IC50, and AC50) were added as binary descriptor, with bit “1” if this activity type was used for measuring the activity value of the compound. The nonbinary properties were scaled to values between 0 and 1, where 1 is equal to the highest value obtained for that particular property from ChEMBL. The compound descriptors for all compounds in the data set are included in Supporting Information file S6.
Protein Descriptors
The protein sequences were transformed into descriptors by dividing every sequence into sections that contained five amino acids per section. From these sections the mean values were calculated for the amino acids’ physicochemical properties: molecular weight, number of chiral atoms, Log D, charge, number of hydrogen bonds and acceptors, rigidity, and number of aromatic bonds. By using the physicochemical properties per five amino acids, the protein descriptors become alignment independent (as we have shown previously) allowing us to expand the application area to diverse multiprotein family data sets.29 Moreover, in this approach descriptor calculation and model training times will be faster compared to the incorporation of physicochemical properties per single amino acid. Additionally, protein taxonomy was taken into account by including classification levels L1, L2, L3, L4, L5, and L6 from ChEMBL.23 These levels were translated into binary values and added as a protein descriptor.
Model Training
The RF models were trained using the ensemble package from the python Scikit-Learn module.35 The amount of trees was set to 500 with random split using seed 12345. Convolutional DNN models were trained to predict binding type and pChEMBL values. The multitask DNN models contained two pyramidal shared hidden layers with 2000 and 100 units, respectively. The DNN implementation was built using Python 2.7 on Keras with Theano36 backend. The following settings were applied: test-size = 0.01 (10%), layer dropout = 0.1, learning rate = 0.0001, momentum = 0.8, patience = 25 s, maximum epochs = 2000, and maximum batch size = 128. The training set was balanced for the binding type classes (orthosteric and allosteric) using oversampling of the underrepresented class. Here, the underrepresented class was multiplied until it reached a corresponding number of entries compared to the opposite class. The number of multiplications was determined by the number of compounds in the majority class divided by the number of compounds in the minority class and rounded off to a whole number. The script to run the RF and DNN models and the configuration file are included as Supporting Information file S7.
Model Validation
Models were validated with a temporal-split approach. The training set consisted of all data from before the year 2013. The test set comprised all data from the publication year 2013 and above. The test set encompassed approximately 15% of the whole class A GPCR data set. The performance of both classification (binding type models) and regression models (pChEMBL prediction models) was evaluated using the metrics MCC, sensitivity, specificity, accuracy, PPV, and NPV.
Glossary
Abbreviations
- DNN
deep neural network
- GPCR
G protein-coupled receptors
- MCC
Matthews correlation coefficient
- NPV
negative predictive value
- PCM
proteochemometrics
- PPV
positive predictive value
- RF
random forest
- RMSE
root-mean-square error
- TM
transmembrane
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jcim.0c00695.
Physicochemical properties of orthosteric and allosteric compounds in ChEMBL per protein family: molecular weight, number of hydrogen bonds, and acceptor (S1) (PDF)
Performance bioactivity models for orthosteric compounds (S2) (XLSX)
Keywords used in text mining (S3) (XLSX)
Pipeline Pilot protocol for text mining (S4) (ZIP)
Python script to run convolutional deep neural networks and random forest models (S7) (ZIP)
Author Contributions
L.B. wrote the manuscript. L.B. and A.v.V. performed the computational experiments. C.C.L. developed the DNN script. L.B., G.J.P.v.W., H.W.T.v.V., and A.P.I.J. contributed to the discussion of the work. All authors have read and approved the final version of the manuscript.
G.J.P.v.W. thanks the Dutch Scientific Council (NWO) and Applied and Engineering Sciences (AES) for funding (VENI #14410).
The authors declare no competing financial interest.
Notes
Compound data set, including binding type annotation (S5), are available at https://data.4tu.nl/articles/Text-mined_orthosteric_and_allosteric_compound_dataset/12717278.
Notes
Compound descriptors (S6) are available at: https://data.4tu.nl/articles/Compound_descriptors_for_text-mined_orthosteric_and_allosteric_dataset/12694739.
Supplementary Material
References
- Soudijn W.; van Wijngaarden I.; IJzerman A. P. Allosteric Modulation of G Protein-Coupled Receptors: Perspectives and Recent Developments. Drug Discovery Today 2004, 9, 752–758. 10.1016/S1359-6446(04)03220-9. [DOI] [PubMed] [Google Scholar]
- Zheng Y.; Qin L.; Zacarías N. V. O.; de Vries H.; Han G. W.; Gustavsson M.; Dabros M.; Zhao C.; Cherney R. J.; Carter P.; Stamos D.; Abagyan R.; Cherezov V.; Stevens R. C.; IJzerman A. P.; Heitman L. H.; Tebben A.; Kufareva I.; Handel T. M. Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists. Nature 2016, 540, 458–461. 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.; Zhang J. Small Molecule Allosteric Modulators of G-Protein-Coupled Receptors - Drug-Target Interactions. J. Med. Chem. 2019, 62, 24. 10.1021/acs.jmedchem.7b01844. [DOI] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The Protein Data Bank. www.rcsb.org. [DOI] [PMC free article] [PubMed]
- Congreve M.; Oswald C.; Marshall F. H. Applying Structure-Based Drug Design Approaches to Allosteric Modulators of GPCRs. Trends Pharmacol. Sci. 2017, 38, 837–847. 10.1016/j.tips.2017.05.010. [DOI] [PubMed] [Google Scholar]
- Wu H.; Wang C.; Gregory K. J.; Han G. W.; Cho H. P.; Xia Y.; Niswender C. M.; Katritch V.; Meiler J.; Cherezov V.; Conn P. J.; Stevens R. C. Structure of a Class C GPCR Metabotropic Glutamate Receptor 1 Bound to an Allosteric Modulator. Science (Washington, DC, U. S.) 2014, 344, 58–64. 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A.; Bellis L. J.; Bento A. P.; Chambers J.; Davies M.; Hersey A.; Light Y.; McGlinchey S.; Michalovich D.; Al-Lazikani B.; Overington J. P. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–1107. 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Westen G. J. P.; Gaulton A.; Overington J. P. Chemical, Target, and Bioactive Properties of Allosteric Modulation. PLoS Comput. Biol. 2014, 10, e1003559. 10.1371/journal.pcbi.1003559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. D.; Lu J.; Carlson H. A. Are There Physicochemical Differences between Allosteric and Competitive Ligands?. PLoS Comput. Biol. 2017, 13, e1005813. 10.1371/journal.pcbi.1005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q.; Zhu Y.; Li J.; Chen Z.; Han G. W.; Kufareva I.; Li T.; Ma L.; Fenalti G.; Li J.; Zhang W.; Xie X.; Yang H.; Jiang H.; Cherezov V.; Liu H.; Stevens R. C.; Zhao Q.; Wu B. Structure of the CCR5 Chemokine Receptor–HIV Entry Inhibitor Maraviroc Complex. Science (Washington, DC, U. S.) 2013, 341, 1387–1390. 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenalti G.; Giguere P. M.; Katritch V.; Huang X.-P.; Thompson A. A.; Cherezov V.; Roth B. L.; Stevens R. C. Molecular Control of δ-Opioid Receptor Signalling. Nature 2014, 506, 191–196. 10.1038/nature12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.; Zhang J.; Gao Z.-G.; Zhang D.; Zhu L.; Han G. W.; Moss S. M.; Paoletta S.; Kiselev E.; Lu W.; Fenalti G.; Zhang W.; Müller C. E.; Yang H.; Jiang H.; Cherezov V.; Katritch V.; Jacobson K. A.; Stevens R. C.; Wu B.; Zhao Q. Structure of the Human P2Y12 Receptor in Complex with an Antithrombotic Drug. Nature 2014, 509, 115–118. 10.1038/nature13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A.; Yano J.; Hirozane Y.; Kefala G.; Gruswitz F.; Snell G.; Lane W.; Ivetac A.; Aertgeerts K.; Nguyen J.; Jennings A.; Okada K. High-Resolution Structure of the Human GPR40 Receptor Bound to Allosteric Agonist TAK-875. Nature 2014, 513, 124–127. 10.1038/nature13494. [DOI] [PubMed] [Google Scholar]
- Oswald C.; Rappas M.; Kean J.; Doré A. S.; Errey J. C.; Bennett K.; Deflorian F.; Christopher J. A.; Jazayeri A.; Mason J. S.; Congreve M.; Cooke R. M.; Marshall F. H. Intracellular Allosteric Antagonism of the CCR9 Receptor. Nature 2016, 540, 462–465. 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- Cheng R. K. Y.; Fiez-Vandal C.; Schlenker O.; Edman K.; Aggeler B.; Brown D. G.; Brown G. A.; Cooke R. M.; Dumelin C. E.; Doré A. S.; Geschwindner S.; Grebner C.; Hermansson N.-O.; Jazayeri A.; Johansson P.; Leong L.; Prihandoko R.; Rappas M.; Soutter H.; Snijder A.; Sundström L.; Tehan B.; Thornton P.; Troast D.; Wiggin G.; Zhukov A.; Marshall F. H.; Dekker N. Structural Insight into Allosteric Modulation of Protease-Activated Receptor 2. Nature 2017, 545, 112–115. 10.1038/nature22309. [DOI] [PubMed] [Google Scholar]
- Weinert T.; Olieric N.; Cheng R.; Brünle S.; James D.; Ozerov D.; Gashi D.; Vera L.; Marsh M.; Jaeger K.; Dworkowski F.; Panepucci E.; Basu S.; Skopintsev P.; Doré A. S.; Geng T.; Cooke R. M.; Liang M.; Prota A. E.; Panneels V.; Nogly P.; Ermler U.; Schertler G.; Hennig M.; Steinmetz M. O.; Wang M.; Standfuss J. Serial Millisecond Crystallography for Routine Room-Temperature Structure Determination at Synchrotrons. Nat. Commun. 2017, 8, 542. 10.1038/s41467-017-00630-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Byrne N.; Wang J.; Bricogne G.; Brown F. K.; Chobanian H. R.; Colletti S. L.; Di Salvo J.; Thomas-Fowlkes B.; Guo Y.; Hall D. L.; Hadix J.; Hastings N. B.; Hermes J. D.; Ho T.; Howard A. D.; Josien H.; Kornienko M.; Lumb K. J.; Miller M. W.; Patel S. B.; Pio B.; Plummer C. W.; Sherborne B. S.; Sheth P.; Souza S.; Tummala S.; Vonrhein C.; Webb M.; Allen S. J.; Johnston J. M.; Weinglass A. B.; Sharma S.; Soisson S. M. Structural Basis for the Cooperative Allosteric Activation of the Free Fatty Acid Receptor GPR40. Nat. Struct. Mol. Biol. 2017, 24, 570–577. 10.1038/nsmb.3417. [DOI] [PubMed] [Google Scholar]
- Liu X.; Ahn S.; Kahsai A. W.; Meng K.-C.; Latorraca N. R.; Pani B.; Venkatakrishnan A. J.; Masoudi A.; Weis W. I.; Dror R. O.; Chen X.; Lefkowitz R. J.; Kobilka B. K. Mechanism of Intracellular Allosteric B2AR Antagonist Revealed by X-Ray Crystal Structure. Nature 2017, 548, 480–484. 10.1038/nature23652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Kim H. R.; Deepak R. N. V. K.; Wang L.; Chung K. Y.; Fan H.; Wei Z.; Zhang C. Orthosteric and Allosteric Action of the C5a Receptor Antagonists. Nat. Struct. Mol. Biol. 2018, 25, 472–481. 10.1038/s41594-018-0067-z. [DOI] [PubMed] [Google Scholar]
- Robertson N.; Rappas M.; Doré A. S.; Brown J.; Bottegoni G.; Koglin M.; Cansfield J.; Jazayeri A.; Cooke R. M.; Marshall F. H. Structure of the Complement C5a Receptor Bound to the Extra-Helical Antagonist NDT9513727. Nature 2018, 553, 111–114. 10.1038/nature25025. [DOI] [PubMed] [Google Scholar]
- La Regina G.; Silvestri R.; Gatti V.; Lavecchia A.; Novellino E.; Befani O.; Turini P.; Agostinelli E. Synthesis, Structure–Activity Relationships and Molecular Modeling Studies of New Indole Inhibitors of Monoamine Oxidases A and B. Bioorg. Med. Chem. 2008, 16, 9729–9740. 10.1016/j.bmc.2008.09.072. [DOI] [PubMed] [Google Scholar]
- Swanson G. T.; Sakai R. Ligands for Ionotropic Glutamate Receptors. Prog. Mol. Subcell. Biol. 2009, 46, 123–157. 10.1007/978-3-540-87895-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento A. P.; Gaulton A.; Hersey A.; Bellis L. J.; Chambers J.; Davies M.; Krüger F. A.; Light Y.; Mak L.; McGlinchey S.; Nowotka M.; Papadatos G.; Santos R.; Overington J. P. The ChEMBL Bioactivity Database: An Update. Nucleic Acids Res. 2014, 42, D1083–90. 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis G. W.; Murcko M. A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39, 2887–2893. 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- Sheridan R. P. Time-Split Cross-Validation as a Method for Estimating the Goodness of Prospective Prediction. J. Chem. Inf. Model. 2013, 53, 783–790. 10.1021/ci400084k. [DOI] [PubMed] [Google Scholar]
- Burggraaff L.; Oranje P.; Gouka R.; van der Pijl P.; Geldof M.; van Vlijmen H. W. T.; IJzerman A. P.; van Westen G. J. P. Identification of Novel Small Molecule Inhibitors for Solute Carrier SGLT1 Using Proteochemometric Modeling. J. Cheminf. 2019, 11, 15. 10.1186/s13321-019-0337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y.; Jing Y.; Wang L.; Ma S.; Jun J. J.; Xie X.-Q. Prediction of Orthosteric and Allosteric Regulations on Cannabinoid Receptors Using Supervised Machine Learning Classifiers. Mol. Pharmaceutics 2019, 16, 2605–2615. 10.1021/acs.molpharmaceut.9b00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.; Wang G.; Li S.; Liu X.; Lu S.; Chen Z.; Song K.; Yan J.; Geng L.; Huang Z.; Huang W.; Chen G.; Zhang J. ASD v3.0: Unraveling Allosteric Regulation with Structural Mechanisms and Biological Networks. Nucleic Acids Res. 2016, 44, D527–D535. 10.1093/nar/gkv902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenselink E. B.; ten Dijke N.; Bongers B.; Papadatos G.; van Vlijmen H. W. T.; Kowalczyk W.; IJzerman A. P.; van Westen G. J. P. Beyond the Hype: Deep Neural Networks Outperform Established Methods Using a ChEMBL Bioactivity Benchmark Set. J. Cheminf. 2017, 9, 45. 10.1186/s13321-017-0232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A.; Hersey A.; Nowotka M.; Bento A. P.; Chambers J.; Mendez D.; Mutowo P.; Atkinson F.; Bellis L. J.; Cibrián-Uhalte E.; Davies M.; Dedman N.; Karlsson A.; Magariños M. P.; Overington J. P.; Papadatos G.; Smit I.; Leach A. R. The ChEMBL Database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A.; Bellis L. J.; Bento A. P.; Chambers J.; Davies M.; Hersey A.; Light Y.; McGlinchey S.; Michalovich D.; Al-Lazikani B.; Overington J. P. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Thiessen P. A.; Bolton E. E.; Chen J.; Fu G.; Gindulyte A.; Han L.; He J.; He S.; Shoemaker B. A.; Wang J.; Yu B.; Zhang J.; Bryant S. H. PubChem Substance and Compound Databases. Nucleic Acids Res. 2016, 44, D1202–D1213. 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIOVIA Pipeline Pilot, Version 16.2.0.58. http://accelrys.com/products/collaborative-science/biovia-pipeline-pilot/.
- Rogers D.; Hahn M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Pedregosa F.; Varoquaux G.; Gramfort A.; Michel V.; Thirion B.; Grisel O.; Blondel M.; Prettenhofer P.; Weiss R.; Dubourg V.; VanderPlas J.; Passos A.; Cournapeau D.; Brucher M.; Perrot M.; Duchesnay E.. Scikit-Learn: Machine Learning in Python. CoRR 2012. [Google Scholar]
- Al-Rfou R.; Alain G.; Almahairi A.; Angermueller C.; Bahdanau D.; Ballas N.; Bastien F.; Bayer J.; Belikov A.; Belopolsky A.; Bengio Y.; Bergeron A.; Zhang Y.; et al. Theano: A Python Framework for Fast Computation of Mathematical Expressions. 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.