Figure 4.
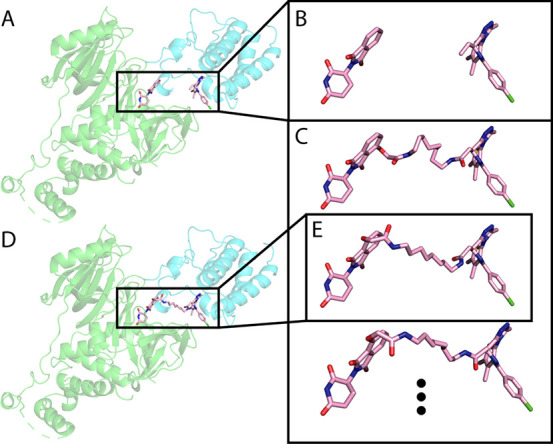
Generating PROTAC conformations in the context of the protein–protein interaction. Using RDKit, we generate up to 100 PROTAC conformations, to match the ligand orientation of each local docking solution. Then, we use Rosetta Packer to choose the best conformation to fit the protein–protein docking model. A. An example local docking solution for 6BOY. B. The ligand orientation, extracted from the local docking solution. C. Constrained conformations that bridge between the ligand orientation. D. Output model of Rosetta after choosing the best constrained conformation. E. The conformation chosen by Rosetta. Green, E3 ligase CRBN; cyan, predicted BRD4; light pink, predicted PROTAC conformation.
