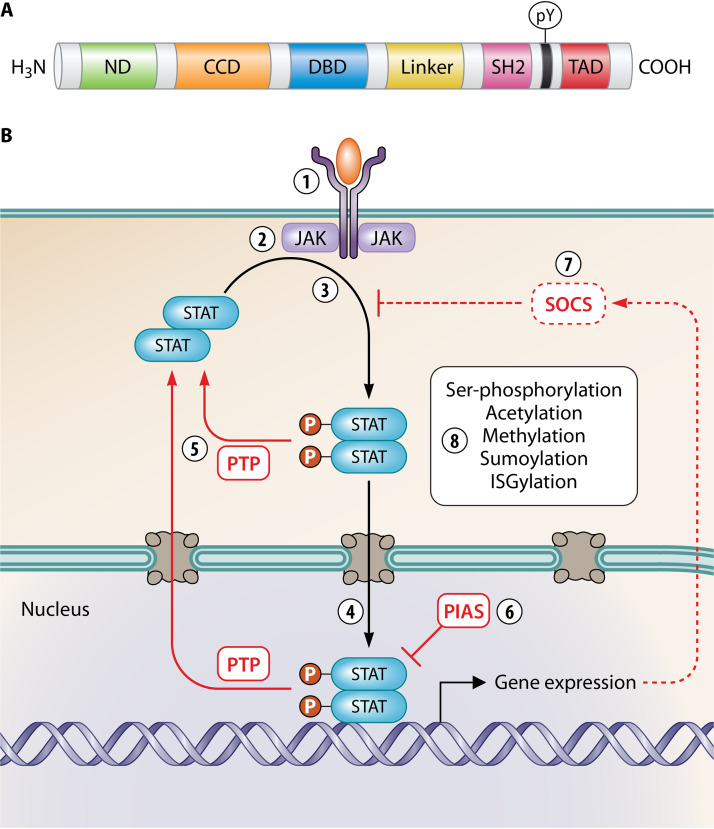
FIG 1.
Structure and signaling pathways of STATs. (A) Schematic representation of the conserved domain structure of STATs: N-terminal domain (ND), coiled-coil domain (CCD), DNA-binding domain (DBD), linker domain, Src homology 2 (SH2) domain, and C-terminal transactivation domain (TAD). The conserved C-terminal tyrosine phosphorylated by JAKs (pY) is indicated. (B) Diverse cytokines signal via a classical JAK-STAT paradigm. (1 and 2) Cytokine, growth factor, etc., bind to the cognate receptor (1), activating specific receptor-associated JAKs (2). (3) JAKs phosphorylate latent antiparallel STAT dimers (P in circles), inducing the formation of parallel dimers through reciprocal pY-SH2 interactions. (4) pY-STAT dimers translocate into the nucleus and bind specific DNA elements, activating gene expression. (5) pY-STATs are dephosphorylated by nuclear and cytoplasmic PTPs to turn off signaling. (6 and 7) STAT signaling is also negatively regulated (blunt arrows) by PIAS proteins (which bind STATs and prevent DNA binding, recruit corepressors, or mediate sumoylation) (6) and SOCS proteins (which are induced by pY-STATs and inhibit JAK kinase activity, bind competitively to STAT-binding sites on receptors, or target cytokine receptors and JAKs for proteasomal degradation; dotted line) (7). Red line, negative regulation; black line, positive regulation. (8) STATs are also positively or negatively regulated by PTMs.