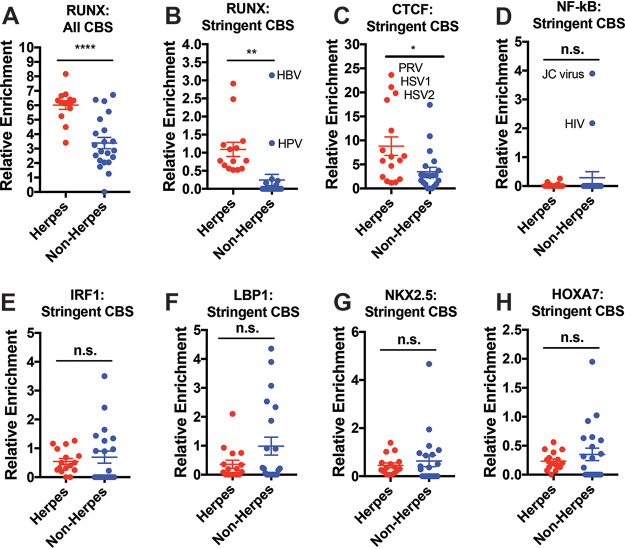
FIG 3.
Herpesviruses are more highly enriched with RUNX CBS, but not other comparable transcription factors. (A) Relative genomic enrichment of all RUNX CBS in herpesviruses and nonherpesviruses. Enrichment calculated as the total CBS/genome size × 10,000. (B) Relative genomic enrichment of RUNX CBS that are located within 2,000 bp of a gene TSS and not present inside a gene body. (C) CBS analysis for CTCF, previously implicated in the latency of HSV-1. (D) CBS analysis for NF-κB, a TF that has already been described to bind to sites within HIV-1 and JC viruses. (E to H) CBS analyses for IRF1, LBP1, NKX2.5, and HOXA7, which have CBS of same length as RUNX (and thus the same expected enrichment). The data are means ± the SE. *, P < 0.05; ***, P < 0.01; *****, P < 0.0001 (versus nonherpesviruses assessed by the Student t test).