Oryctes rhinoceros nudivirus has been an effective biocontrol agent against the coconut rhinoceros beetle in Southeast Asia and the Pacific Islands for decades. The recent outbreak of these beetles in many South Pacific islands has had a significant impact on livelihoods in the region. It has been suggested that the resurgence and spread of the pest are related to the presence of low-virulence isolates of OrNV or virus-tolerant haplotypes of beetles. We examined viral genomic and transcriptional variations in chronically infected beetles from different geographical populations. A high number of polymorphic sites among several geographical strains of OrNV were identified, but potentially only a few of these variations in the genome are involved in functional changes and can potentially alter the typical function. These findings provide valuable resources for future studies to improve our understanding of the OrNV genetic variations in different geographic regions and their potential link to virus pathogenicity.
KEYWORDS: Oryctes rhinoceros nudivirus (OrNV), coconut rhinoceros beetle, South Pacific Islands, biological control, genomic variation, viral transcriptome, OrNV, Oryctes rhinoceros nudivirus
ABSTRACT
Oryctes rhinoceros nudivirus (OrNV) is a double-stranded DNA (dsDNA) virus which has been used as a biocontrol agent to suppress the coconut rhinoceros beetle (Oryctes rhinoceros) in Southeast Asia and the Pacific Islands. A new wave of O. rhinoceros incursions in Oceania is thought to be related to the presence of low-virulence isolates of OrNV or virus-tolerant haplotypes of beetles. In this study, chronically infected beetles were collected from Philippines, Fiji, Papua New Guinea (PNG), and the Solomon Islands (SI). RNA sequencing (RNA-seq) was performed to investigate the global viral gene expression profiles and for comparative genomic analysis of structural variations. Maximum likelihood phylogenic analysis indicated that OrNV strains from the SI and Philippines are closely related, while OrNV strains from PNG and Fiji formed a distinct adjacent clade. We detected several polymorphic sites with a frequency higher than 35% in 892 positions of the viral genome. Nonsynonymous mutations were detected in several hypothetical proteins and 15 nudivirus core genes, such as gp034, lef-8, lef-4, and vp91. We found limited evidence of variation in viral gene expression among geographic populations. Only a few genes, such as gp01, gp022, and gp107, were differentially expressed among different strains. Additionally, small RNA sequencing from the SI population suggested that OrNV is targeted by the host RNA interference (RNAi) response with abundant 21-nucleotide small RNAs. Some of these genomic changes are specific to the geographic population and could be related to particular phenotypic characteristics of the strain, such as viral pathogenicity or transmissibility, and this requires further investigation.
IMPORTANCE Oryctes rhinoceros nudivirus has been an effective biocontrol agent against the coconut rhinoceros beetle in Southeast Asia and the Pacific Islands for decades. The recent outbreak of these beetles in many South Pacific islands has had a significant impact on livelihoods in the region. It has been suggested that the resurgence and spread of the pest are related to the presence of low-virulence isolates of OrNV or virus-tolerant haplotypes of beetles. We examined viral genomic and transcriptional variations in chronically infected beetles from different geographical populations. A high number of polymorphic sites among several geographical strains of OrNV were identified, but potentially only a few of these variations in the genome are involved in functional changes and can potentially alter the typical function. These findings provide valuable resources for future studies to improve our understanding of the OrNV genetic variations in different geographic regions and their potential link to virus pathogenicity.
INTRODUCTION
Oryctes rhinoceros nudivirus (OrNV) (genus Alphanudivirus) belongs to the diverse Nudiviridae family of rod-shaped viruses with large single covalently closed circular double-stranded DNA (dsDNA) genomes, and it replicates in the nucleus of the coconut rhinoceros beetle (Oryctes rhinoceros). Phylogenetically, nudiviruses are related to the baculoviruses but form a separate lineage and share a set of about 21 “core genes” which are involved in host-virus interaction, replication, and metabolism (1, 2). Nudiviruses were previously considered to be nonoccluded baculoviruses based on their ultrastructure, but later it was removed from the family Baculoviridae due to absence of occlusion bodies (2, 3). Nudiviruses are closely related to other large DNA viruses such as the insect-specific salivary gland hypertrophy virus (SGHV) and white spot syndrome virus (WSSV), which infect crustaceans such as penaeid shrimp. The typical pathologies observed in nudivirus-infected insects and crustaceans suggest lethal infections in larvae and chronic disease in adults of various terrestrial and aquatic species (2, 3).
The genome size of OrNV ranges between 125 and 127 kbp, and the genome includes 130 to 140 open reading frames (ORFs) in different sequenced isolates (4, 5), making it significantly smaller than the closely related Heliothis zea nudivirus (HzNV) (228 to 230 kbp). The identified open reading frame in OrNV encodes proteins in the range of ∼5 kDa (51 amino acids [aa]) to ∼140 kDa (1,280 aa) (4). The synteny and structure of homologs between nudivirus genomes are poorly conserved. OrNV, HzNV, and Gryllus bimaculatus nudivirus (GbNV) were the first described insect nudiviruses (described in the 1960s), and they have 33 open reading frames (ORFs) in common (1). These 33 genes include 32 “core genes” which are present in all nudivirus genomes and of which 21 are homologs of baculovirus genes (6).
The coconut rhinoceros beetle (Oryctes rhinoceros), which is considered a globally invasive species (7), is a pest of palm trees and a severe threat to livelihoods in tropical Asia and the Pacific Islands. Oryctes rhinoceros is indigenous to Southeast Asia, and OrNV was initially discovered and isolated from insects in Malaysia in 1963, providing a biological control agent for O. rhinoceros that has been successfully used across the region for 50 years (8).
New incursions of the pest into previously O. rhinoceros-free countries and territories have been reported in recent years, beginning in Guam in 2007, followed by Hawaii (2013), the Solomon Islands (2015), and, most recently, Vanuatu and New Caledonia in 2019 (9). It has been suggested that the resurgence and spread of the pest are related to an OrNV-resistant haplotype (CRB-G), which is identified by single nucleotide polymorphisms (SNPs) in cytochrome c oxidase subunit I (CoxI) (10). Since the release of OrNV into the Pacific, there have been some reports that indicate possible failures of the biocontrol agent, including repeated outbreaks of the beetle, the inconsistency of mortality caused by OrNV, and the lack of disease symptoms or low incidence of the virus in wild populations (11). Accordingly, the presence of low-virulence isolates of virus or tolerate haplotypes of beetles in different geographical regions has been proposed (12, 13). Several genomic variants of OrNV have been reported across the region based on endonuclease restriction analyses (14, 15). Still, the actual genetic diversity present within the wild-type populations of OrNV remains unknown. Crawford and Zelazny detected the first viral structural variations 4 years after the initial release of different isolates of the virus in the Maldives. Using a restriction endonuclease digestion approach, they described some genomic alterations due to insertion and point mutations and also documented recombination between two of the strains of the virus released, suggesting rapid evolution of the virus (14).
Geographical populations of the virus may vary in virulence due to their genetic diversity. It has been reported that the failure of the virus to maintain control of O. rhinoceros in Malaysia may be due to low virulence of endemic OrNV strains (15–17). The successful establishment of OrNV in O. rhinoceros populations is also critically dependent on virus transmissibility, and this too is likely to have a genetic basis. Different levels of transmission have been quantified for OrNV across the region. Still, too little information is available to understand the genetic basis of differences between the virulence and transmissibility of different geographical OrNV strains. Releasing several strains of the virus, as an essential component of pest management strategies, potentially intensifies the genetic diversity of OrNV in the region.
Previous studies have found widespread genomic variations in some large DNA viruses, like baculoviruses (18, 19). Many comparative analyses performed on baculovirus-host interaction models suggest that the differences in virulence and transmissibility involve multiple molecular pathways. Nonetheless, some mutations in key genes can be responsible for low transmissibility or virulence in a particular phenotype of the virus (20, 21). A single wild-type nucleopolyhedrovirus (NPV)-infected Panolis flammea caterpillar was found to contain 24 genotypic variants based on restriction fragment length polymorphism (22). However, the fitness cost associated with genetic diversity in the population and whether this genetic diversity can be preserved in the population are unclear. Some nonsynonymous mutations in core genes might have a significant impact on the virus fitness within different populations (19). However, structural variants with low frequency can also be maintained within the population regardless of their fitness cost and can be a dominant genotype when suitable conditions arise (19).
The successful infection of the host gut tissue by OrNV particles is a crucial step that determines the success of the viral infection in O. rhinoceros. Many host and viral genes are involved in this critical event, as the midgut represents the first line of cellular defense against viral infection. In baculoviruses, a complex of proteins called per os infectivity factors (PIFs) mediate the binding and entry of virus particles into midgut epithelial cells. Undoubtedly, successful nudivirus infection involves a very complicated and coordinated expression of all viral genes. Any transcriptional variation or nonsynonymous mutations of viral genes which disable the normal function of these proteins could reduce virus virulence and interrupt vertical or horizontal transmission of the virus.
Unfortunately, the lack of information on the global gene expression pattern of OrNV in persistently (or chronically) infected insect hosts or even in cultured insect cells prevents us from having a clear view of host-pathogen interactions at the viral gene transcriptional level. We recently detected an extremely high prevalence of viral infection in O. rhinoceros adult specimens across the region, even in the host haplotype (CRB-G) believed to be resistant to OrNV (9). Here, we present the first genome-wide study of the genomic diversity of OrNV strains across the Pacific. This study generated a compendium of OrNV genomic variants and gene expression profiles in different geographical strains which infect the different mitochondrial lineages (CRB-G, CRB-S, and CRB-PNG) of O. rhinoceros. Furthermore, some of these genomic changes are specific to different geographic populations and could be related to particular phenotypic characteristics of the strain, such as viral pathogenicity or transmissibility.
RESULTS AND DISCUSSION
Evidence of codivergence of OrNV strains with their O. rhinoceros hosts.
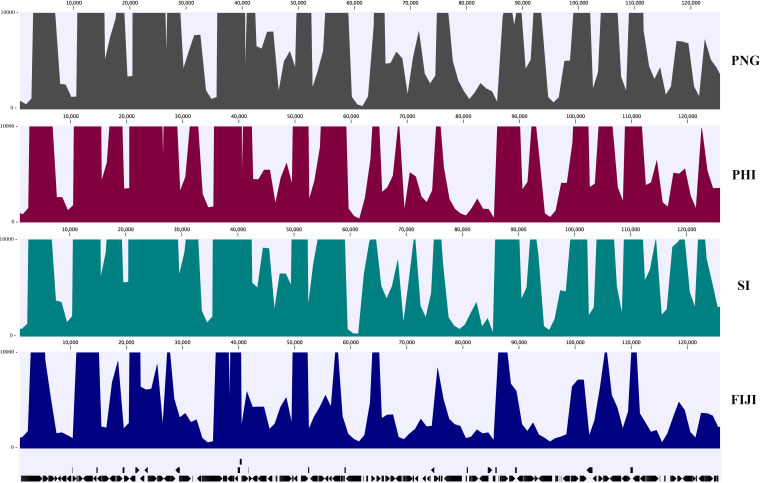
In this study, O. rhinoceros adults were collected from Philippines in their native region and from Fiji (Sigatoka, Viti Levu), Papua New Guinea (PNG; Kimbe, New Britain), and the recently invaded Solomon Islands (Honiara, Guadalcanal) in non-native regions in the South Pacific Islands (Fig. 1A). The total RNAs from adults O. rhinoceros infected with wild-type OrNV were sequenced, and in total, 42,678,016 Illumina paired-end reads were mapped to the OrNV genome. The proportion of reads mapped to the virus genome in each library was between 0.54% and 18.92% of the total reads (Table 1). We used the recently sequenced genome (MN623374) of a Solomon Islands strain of OrNV as a reference in this study (5), and the viral genome consensus sequences have been generated for each library (Fig. 2).
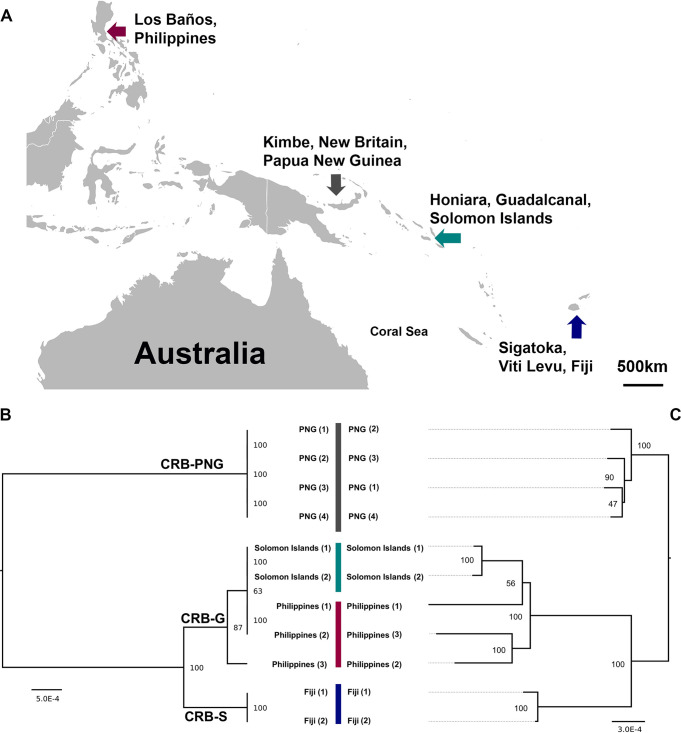
FIG 1.
(A) Geographic locations of coconut rhinoceros beetles (O. rhinoceros) chronically infected with OrNV from Southeast Asia and the Pacific Islands. (B) Host mitochondrial maximum likelihood phylogeny based on O. rhinoceros CoxI gene sequence. Four different haplotypes of O. rhinoceros in three major mitochondrial lineages (CRB-PNG, CRB-S and CRB-G) were identified. (C) Maximum likelihood phylogeny analysis of OrNV strains. Samples from the Solomon Islands and Philippines are closely related to each other, while individuals from PNG and Fiji are separated from them and located in another clade. Harmonious topologies of both host and OrNV strains provide evidence of codivergence of OrNV strains with their O. rhinoceros hosts. Branch length indicates nucleotide substitutions per site; trees were bootstrapped 1,000 times. The host phylogenetic tree is midpoint rooted.
TABLE 1.
Distribution of viral reads among different RNA-seq libraries
| Population | Sample identifier | No. of reads mapped to viral genome | Viral reads in library (%) | Gene (%) | Intergenic (%) |
|---|---|---|---|---|---|
| Fiji | FIJI 1 | 465,294 | 0.91 | 94.58 | 5.42 |
| FIJI 2 | 2,498,061 | 6.34 | 93.63 | 6.37 | |
| Philippines | PHI 1 | 8,188,367 | 18.92 | 94.08 | 5.92 |
| PHI 2 | 6,129,400 | 13.22 | 93.69 | 6.31 | |
| PHI 3 | 525,590 | 0.93 | 92.88 | 7.12 | |
| PNG | PNG 1 | 1,361,190 | 3.21 | 93.31 | 6.69 |
| PNG 2 | 5,421,019 | 12.65 | 93.58 | 6.42 | |
| PNG 3 | 4,919,054 | 11.87 | 94.1 | 5.9 | |
| PNG 4 | 5,516,617 | 13.62 | 93.28 | 6.72 | |
| Solomon Islands | SI 1 | 7,406,766 | 14.33 | 94.04 | 5.96 |
| SI 2 | 246,658 | 0.54 | 91.54 | 8.46 |
FIG 2.
Average coverage of RNA-seq data mapped to the OrNV genome (Solomon Islands [SI] strain). A consensus sequence of virus was generated for each sample. PHI, Philippines.
Maximum likelihood phylogeny analysis of four different wild-type populations of OrNV based on their complete genome sequence consensus revealed two major clades. These data show that samples from Solomon Islands and Philippines are closely related to each other, while samples from PNG and Fiji are separated from them and located in another clade (Fig. 1). The phylogenic analysis of field-collected O. rhinoceros based on the host partial CoxI gene revealed three major mitochondrial lineages, CRB-G, CRB-S, and CRB-PNG. All O. rhinoceros individuals collected from PNG (Kimbe, New Britain) and Fiji belonged to the mitochondrial lineages CRB-PNG and CRB-S, respectively. The individuals from Philippines and the recently invaded Solomon Islands belonged to the CRB-G clade (Fig. 1C). The virus strains from the Solomon Islands and Philippines also belonged to one major clade (Fig. 1B). Harmonious topologies of both host and OrNV strains provide evidence of codivergence of OrNV strains with their O. rhinoceros hosts. While bootstrap values for most clades of the CoxI genes and OrNV phylogenetic trees showed strong support for geographically distinct clades, there was lower support between the OrNV Solomon Islands 1-2 clade and the Philippines 1-2 sample clade. This lower bootstrap support may be due to inadequate sampling of OrNV or evidence for recombination between the Solomon Islands and Philippines samples, as OrNV is both known to have undergone recombination (14) and to be prone to large genome inversions (5). While we did not detect evidence for recombination within the 196 codon-aligned genes in OrNV using GENECOV and Bootscan analysis in RDP4, multigene and structural changes in OrNV genomes are unlikely to be resolved through RNA sequencing (RNA-seq) alone. Therefore, further sampling, DNA sequencing, and de novo assembly of OrNV strains may be necessary for further examination of recombination between these geographic isolates and clarification of these nodes.
Previously, Marshall et al. (10) reported that the natural infection rate of OrNV ranges from 45.2% to 64.5% in O. rhinoceros from a wide range of countries in Southeast Asia and the Pacific. However, they could not detect the presence of the OrNV in specimens from the Solomon Islands, Philippines, and Oahu, which were believed to be the CRB-G haplotype. The source of the virus found in our 2019 collection from the Solomon Islands is not known (5); the virus could have been introduced deliberately as a biological control agent or along with an incursion of infected beetles from neighboring islands (CRB-PNG from PNG) (9). However, due to the high level of similarity and lack of any evidence that OrNV (Philippines strain) was introduced into the country as a biological control agent, our work suggests that the OrNV came to the Solomon Islands with an incursion of infected CRB-G beetles that originated in Southeast Asia.
Whole-genome structural variant analysis of OrNV strains.
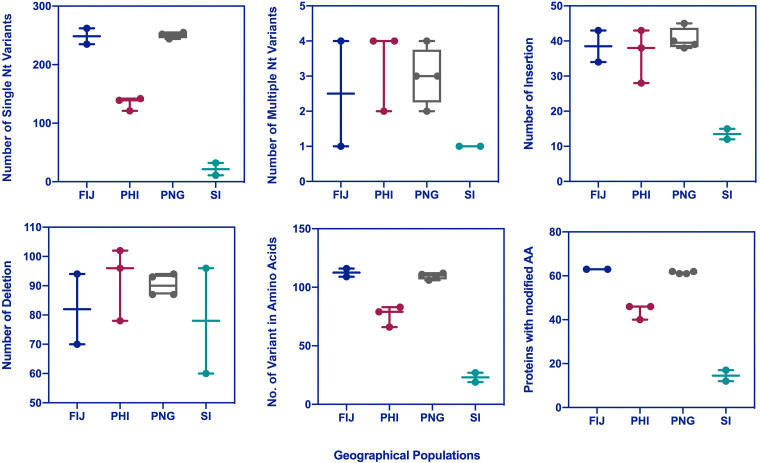
We detected several SNPs with a frequency higher than 35% in 892 positions of the 125,917-bp viral genome sequence, which were found in pair-end reads positioned in both orientations of the genome. In two samples collected from the Solomon Islands, 87 to 141 variants were identified in which deletion was their major variation compared to the newly sequenced genome reference (MN623374). The maximum number of genomic variants such as single nucleotide variants (SNV), insertion, and deletion and the maximum number of variants in amino acid sequence (nonsynonymous mutations) were found in individuals from Fiji and PNG (Fig. 3). We detected 12 to 17 putative proteins with amino acid modifications in samples collected from the Solomon Islands, while the number of such proteins reached 61 to 63 in samples from Fiji and PNG. OrNV in Philippines showed more similarity to OrNV in individuals from the Solomon Islands based on the number of identified SNV and nonsynonymous modifications. We also mapped the viral reads to a previously sequenced genome of a Malaysian OrNV isolate (Ma07). We found between 475 and 532 polymorphic sites between our different viral consensus sequences with the Malaysian isolate (Table 2). These data revealed more similarity between Ma07 and samples collected from PNG and Fiji.
FIG 3.
Genomic variants of different geographical populations of OrNV compared to the newly sequenced reference genome of a Solomon Islands strain. Single nucleotide variants (SNV), multiple nucleotide variants (MNV), insertion, deletion, the number of variants in amino acids (AA; nonsynonymous mutations), and OrNV gene products with modified amino acids are indicated. The maximum numbers of genomic variants were found in individuals from Fiji and PNG. As expected, less variation was found in samples collected from the Solomon Islands.
TABLE 2.
Pairwise comparison of the whole-genome sequence between Malaysian isolate of OrNV (Ma07) and consensus sequences of other geographical populations of OrNV
| Sample identifier | SI 1 | SI 2 | PHI 1 | PHI 2 | PHI 3 | PNG 1 | PNG 2 | PNG 3 | PNG 4 | Fiji 1 | Fiji 2 | Ma 07 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SI 1 | 96a | 382 | 453 | 405 | 508 | 513 | 506 | 492 | 531 | 534 | 513 | |
| SI 2 | 99.92b | 394 | 449 | 367 | 492 | 515 | 509 | 501 | 496 | 528 | 475 | |
| PHI 1 | 99.70 | 99.69 | 482 | 309 | 527 | 526 | 515 | 563 | 546 | 546 | 527 | |
| PHI 2 | 99.65 | 99.65 | 99.62 | 484 | 561 | 580 | 588 | 640 | 640 | 614 | 550 | |
| PHI 3 | 99.68 | 99.71 | 99.76 | 99.62 | 540 | 517 | 522 | 560 | 501 | 533 | 482 | |
| PNG 1 | 99.60 | 99.61 | 99.59 | 99.56 | 99.58 | 132 | 158 | 194 | 424 | 419 | 449 | |
| PNG 2 | 99.60 | 99.60 | 99.59 | 99.55 | 99.60 | 99.90 | 103 | 178 | 403 | 384 | 478 | |
| PNG 3 | 99.60 | 99.60 | 99.60 | 99.54 | 99.59 | 99.88 | 99.92 | 174 | 396 | 374 | 478 | |
| PNG 4 | 99.61 | 99.61 | 99.56 | 99.50 | 99.56 | 99.85 | 99.86 | 99.86 | 449 | 446 | 532 | |
| FIJI 1 | 99.58 | 99.61 | 99.57 | 99.50 | 99.61 | 99.67 | 99.68 | 99.69 | 99.65 | 104 | 475 | |
| FIJI 2 | 99.58 | 99.59 | 99.57 | 99.52 | 99.58 | 99.67 | 99.70 | 99.71 | 99.65 | 99.92 | 479 | |
| Ma 07 | 99.60 | 99.63 | 99.59 | 99.57 | 99.62 | 99.65 | 99.63 | 99.63 | 99.58 | 99.63 | 99.62 |
Upper comparisons represent the numbers of single nucleotide differences between isolates.
Lower comparisons are the percentages of similarity between individual isolates.
Genomic variants have been found in both coding and noncoding regions of the OrNV genome. Generally, mutations in coding regions are likely to have a significant impact on the genome, whereas in noncoding regions, such mutations are more likely to be neutral. However, certain noncoding regions of baculoviruses could be important components of the viral genomes (20).
Nonsynonymous mutations which do result in amino acid modification and could potentially interrupt protein function were detected in 15 nudivirus core genes and several hypothetical proteins (Table 3; see also Table S1 in the supplemental material). We found up to four amino acid modifications in DNA helicase (GenBank accession no. QHG11272.1) among individuals from different populations. A nonsynonymous mutation (c.5C>T) in OrNV_gp034 caused an amino acid modification in DNA helicase in an individual sample from the Solomon Islands. This amino acid substitution from threonine to methionine can also change the biochemical properties (polarity changed from neutral polar to nonpolar) of this critical protein. Therefore, it might affect protein function and possibly virus performance. Other nonsynonymous mutations in this gene do not change the polarity of the amino acid residue. One of these amino acid changes (p.Leu211Phe) is specific to samples isolated from Fiji. It has been suggested that mutations in the genes involved in DNA replication might affect the rate of virion production in baculoviruses (e.g., Spodoptera exigua multiple nucleopolyhedrovirus [SeMNPV]). It thus might alter the spread of the infection within the host (20). DNA helicase is one of the most rapidly evolving genes in Drosophila innubila nudivirus (DiNV) and Kallithea virus and may be essential for adaptation to a new host system (23, 24).
TABLE 3.
Number of amino acid modifications in OrNV core gene products
| Gene identifier | Gene | Gene product | Protein NCBI accession no. | Value for: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fiji 1 | Fiji 2 | PHI 1 | PHI 2 | PHI 3 | PNG 1 | PNG 2 | PNG 3 | PNG 4 | SI 1 | SI 2 | ||||
| DNA replication | ||||||||||||||
| OrNV_gp001 | dnapol | DNA polymerase B | QHG11240.1 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| OrNV_gp034 | helicase | DNA helicase | QHG11272.1 | 4 | 4 | 3 | 1 | 2 | 3 | 3 | 3 | 3 | ||
| OrNV_gp108 | helicase | DNA helicase 2 | QHG11340.1 | 1 | 1 | |||||||||
| Transcription: | ||||||||||||||
| OrNV_gp020 | p47 | p47 protein | QHG11259.1 | |||||||||||
| OrNV_gp064 | lef-8 | Late expression factor 8 | QHG11301.1 | 1 | 1 | 1 | ||||||||
| OrNV_gp096 | lef-9 | Late expression factor 9 | QHG11328.1 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | ||
| OrNV_gp042 | lef-4 | Late expression factor 4 | QHG11280.1 | 1 | 1 | |||||||||
| OrNV_gp052 | lef-5 | Late expression factor 5 | QHG11289.1 | |||||||||||
| OrNV_gp059 | lef-3 | Late expression factor 3 | QHG11296.1 | |||||||||||
| Envelope component: | ||||||||||||||
| OrNV_gp017 | pif-2 | Per os infectivity 2 | QHG11256.1 | 1 | ||||||||||
| OrNV_gp060 | pif-1 | Per os infectivity 1 | QHG11297.1 | 2 | 1 | 1 | 1 | 1 | ||||||
| OrNV_gp107 | pif-3 | Per os infectivity 3 | QHG11339.1 | 1 | ||||||||||
| OrNV_gp033 | pif-4 | 19-kDa protein | QHG11271.1 | |||||||||||
| OrNV_gp115 | pif-5 | ODV-E56 protein | QHG11347.1 | 1 | ||||||||||
| OrNV_gp126 | p74 | p74 protein | QHG11358.1 | 3 | 3 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| OrNV_gp004 | ac81 | Ac81-like protein | QHG11243.1 | 1 | 1 | |||||||||
| OrNV_gp113 | p33 | Ac92-like protein | QHG11345.1 | |||||||||||
| Capsid component: | ||||||||||||||
| OrNV_gp087 | 38K | 38K protein | QHG11320.1 | |||||||||||
| OrNV_gp072 | ac68 | Ac68-like protein | QHG11307.1 | |||||||||||
| OrNV_gp030 | vlf-1 | Very late expression factor 1 | QHG11269.1 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | ||
| OrNV_gp022 | gp72 | GrBNV_gp72-like protein | QHG11261.1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| OrNV_gp015 | vp39 | Viral capsid-associated protein 39 | QHG11254.1 | 1 | ||||||||||
| OrNV_gp106 | vp91 | Viral capsid-associated protein 91 | QHG11338.1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | ||||
We identified nonsynonymous mutations in several genes which are potentially involved in transcription, including genes for three subunits of the RNA polymerase (lef-4, lef-8, and lef-9) (Table 3 and Table S1). Interestingly, a similar event has been reported for Autographa californica multiple nucleopolyhedrovirus (AcMNPV) (19) and SeMNPV (20). In this study, amino acid variants in lef-9 did not change protein polarity. Still, nonsynonymous mutations in lef-8 and lef-4 had an impact on biochemical properties of the protein in some individuals from Fiji and PNG. OrNV_gp64, which encodes lef-8 protein, was also overexpressed in OrNV collected from Fiji (fold change, 4.38; false-discovery rate [FDR], 0.048), and the lowest expression value was detected in individuals from Philippines. lef-4 is an RNA capping enzyme and was originally identified as being essential for late transcription. A deletion mutant in the Bomby mori nucleopolyhedrovirus (BmNPV) lef-8 homolog (Bm39) was not viable, and inactivation of BmNPV lef-8 prevented virus replication (25, 26). Theze et al. (20) also suggested that mutations in genes for the subunits of the viral RNA polymerase, lef-9 and p47 ORFs, are associated with vertical transmission and pathogenicity. We did not detect any mutations in OrNV_gp020, which encodes p47 protein, in individuals from across the Pacific, suggesting that this gene is essential for OrNV replication. However, it is challenging to definitively link specific mutations to virus performance because virulence and virus performance are derived from the combination of the genetic material present at the whole-genome scale.
Genes involved in baculovirus primary infection encoding essential components of the per os infectivity complex, such as pif-1, pif-2, and pif-3, also have some nonsynonymous mutations in their sequences across the different geographical population of OrNV (Table 3 and Table S1). Although we do not have experimental data about the function of these genes in nudiviruses, the products of pif genes, PIF proteins, are conserved across the baculoviruses and nudiviruses. These proteins bind to insect midgut cells and potentially are essential for infection. We detected only one amino acid modification in PIF-3 and PIF-2 in one of the samples collected from the Philippines. There is a conserved amino acid change (p.Val184Ile) in PIF-1 of all individuals from the PNG population, but it has no impact on protein polarity. Chateigner et al. (19) suggested that the presence of different variants of the PIF proteins in the AcMNPV might allow the binding of these proteins to a wider range of host cell, which could be advantageous for establishment of viral infection.
OrNV_gp126, which encodes p74 protein, is another gene with a role in per os infectivity in baculoviruses, and we detected amino acid changes in all individuals. This protein is critical in host-pathogen interactions in the insect midgut, and its deletion prevents the transmission of recombinant virus in insects, but the mutation does not affect replication in cultured cells (27). Two of the amino acid changes were specific to the Fiji population and altered protein polarity.
The very late factor (VLF) appears to be a structural, conserved protein that is present in both baculoviruses (BV and ODV) and nudiviruses and which is required for the production of nucleocapsids. Several deletion/insertions of serine (p349̂350) and glutamine (p644̂645) have been detected in vlf-1 in all the OrNV samples; this could be due to a sequencing error (homopolymer stretches) or the presence of actual mutation sites (Table 3 and Table S1). We also detected two mutation sites in OrNV_gp106, which encodes VP91 protein. One of those mutations (p.Glu564Asp) is specific to OrNV from the Solomon Islands and Philippines, and the other (p.Ala117Thr) is specific to OrNV from Fiji. Changing alanine to threonine also alters protein polarity from nonpolar to polar, and it could cause critical conformational changes affecting protein structure. Theze et al. showed that some variants of vp91 in SeMNPV could be associated with pathogenicity (20).
Limited evidence of OrNV gene expression variation between chronically infected populations.
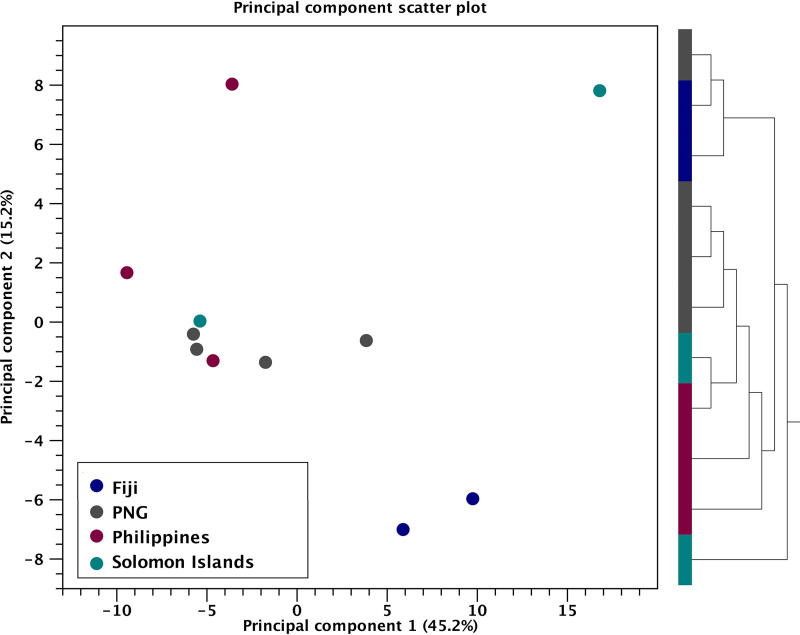
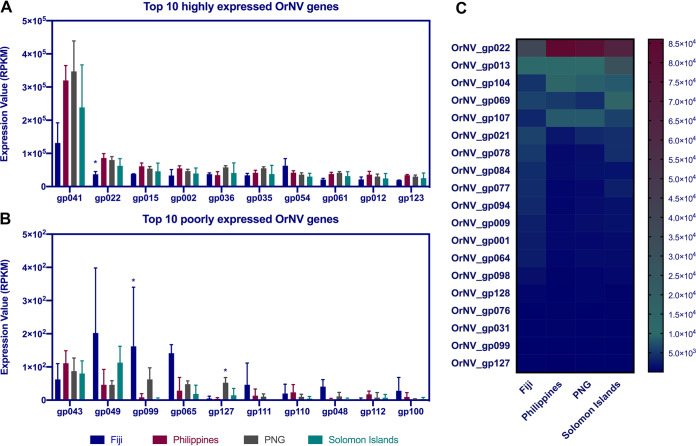
Principal-component analysis of OrNV samples from different countries based on overall viral gene expression profiles can differentiate each of the OrNV populations. Two samples from Fiji had similar profiles, and their expression pattern was slightly different from those of the other populations (Fig. 4). Two individuals from the Solomon Islands and Philippines are located outside their population cluster, probably due to varying stages of infection, specific gene expression patterns, and low viral reads in those libraries (Fig. 4 and Table 1). However, we performed library size normalization using the TMM (trimmed mean of M values) method, and library sizes were then used as part of the presample normalization. To get a better view of the gene expression profile in each sample, we ranked the genes based on their value for reads per kilobase per million (RPKM) and filtered the most highly and poorly expressed gene in each library.
FIG 4.
Principal-component analysis of OrNV samples from different regions based on the global viral gene expression profile. The plot shows the projection of the samples onto the two-dimensional space spanned by the first and second principal components of the covariance matrix. The expression levels used as input are normalized log CPM (counts per million reads mapped) values. The hierarchical clustering clusters values. The hierarchical clustering clusters each individual population by the similarity of their expression profiles over the set of samples and the complete linkage calculated by Pearson’s correlation coefficient.
OrNV_gp41 was the most highly expressed gene with an unknown function in all individual libraries. This 342-bp nudivirus-specific gene shows significant similarity to GrBNV_gp95 and does not have a known homolog in baculoviruses. Further sequence analysis using InterProScan and the GO database showed that some parts of OrNV_gp41 gene product are integral components of the hydrophobic region of the membrane (GO:0016021). This virus fragment also integrated into the brown planthopper (Nilaparvata lugens) genome (28). Recently, it has been identified in endogenous nudivirus-like viruses of parasitoid wasps, i.e., Cotesia congregata bracovirus (CcBV) and Chelonus inanitus bracovirus (CiBV), and its expansion into a gene family in a wasp genome also reported from Venturia canescens virus-like particles (VcVLP) (29).
OrNV_gp022 (GrBNV_gp72-like) is a member of alphanudivirus core genes with unknown function, which is in the top 10 highly expressed genes in all OrNV populations. However, its normalized expression value in the Fiji population is significantly lower than in other samples (Fig. 5 and Table S2). Other genes among the top 10 most highly expressed genes are OrNV_gp015, gp002, gp054, gp012, and gp023, which encode viral capsid Vp39, a trypsin-like serine protease, glycoprotein gp83-like, ODV-E66, and guanylate kinase-like proteins, respectively.
FIG 5.
(A and B) Top 10 highly and poorly expressed OrNV genes in different geographical strains. (C) Only 19 genes are differentially expressed among four OrNV strains. Each column in the heat map corresponds to a geographical region and each row corresponds to a differentially expressed viral gene. TMM normalization was used to make samples comparable, and then a z-score normalization was applied to make the gene expression values comparable.
Trypsin-like serine proteases are nonstructural proteins with a conserved domain (cl21584) which are found in several RNA and DNA viruses and cellular organisms (30). These proteins are essential for virus maturation (31), and they are involved in proteolysis through serine-type endopeptidase activity. Posttranslational modifications of these enzymes play a crucial role in virus replication (32). Previously, it has been shown that trypsin-serine proteases have a ratio of nonsynonymous to synonymous evolutionary changes (dN/dS ratio) above 0.5 between DiNV and OrNV, which indicate that this gene is under purifying selection (dN/dS ratio < 1) and unconstrained evolution or putative adaptation (dN/dS ratio > 0.5) (23). However, we did not detect any amino acid modification in this gene within the different OrNV populations.
Recently, it has been experimentally demonstrated that a nudivirus-encoded protein, gp83, inhibits the Toll signaling pathway in the fruit fly. Drosophila melanogaster Kallithea virus (KV), which is a new member of the family Nudiviridae, and DiNV suppress host immune responses by regulating NF-κB transcription factors (33). This immunosuppressive activity of gp83 against D. melanogaster NF-κB signaling is conserved, and probably gp83 shows similar activity in other members of Nudiviridae. This gene is the second most highly expressed gene in samples collected from Fiji, and its normalized expression value is slightly higher than in other populations (Fig. 5).
ODV-E66 protein, which has a chondroitin AC/alginate lyase (IPR008929) domain, is potentially involved in viral envelope formation and facilitates viral protein movement during infection by degrading larval peritrophic membrane in baculoviruses (34). This protein has been reported from baculoviruses and other nudiviruses (such as Homarus gammarus nudivirus and Penaeus monodon nudivirus), and it has been suggested that ODV-E66 is a per os infectivity factor (3, 35). Although odv-e66 is not among the top 20 most highly expressed genes in AcMNPV, its expression pattern has a strong negative correlation between the Trichoplusia ni midgut and a T. ni cell line (Tnms42). Overexpression in midgut tissue confirms its responsibility for the trafficking of viral proteins during infection (21).
OrNV_gp107, which encodes PIF-3, is highly expressed in OrNV strains from Philippines, and its expression is 3.56 times higher than in samples collected from Fiji (Fig. 5C). Although expression of pif genes is necessary for successful infection of gut tissue, it has been shown that overexpression of pif-1 in Spodoptera frugiperda NPV is unfavorable for viral population fitness and can cause more mutation in this gene and development of viral genotypes with lack of pif-1 expression capabilities (36). There are several pif genes in OrNV which encode essential components of envelope proteins and encode an N-terminal hydrophobic sequence in combination with several adjacent positively charged amino acids.
Most of the OrNV poorly expressed genes encode hypothetical proteins, and we do not have enough information about their potential role in the host-pathogen interaction. OrNV_gp99, which encodes Ac120-like protein, is one the most poorly expressed genes in OrNV but their expression was significantly upregulated in samples collected from Fiji and almost undetectable in Philippines and Solomon Islands populations (Fig. 5 and Table S2). We recently found a 6-nucleotide (nt) substitution in OrNV_gp99 when we compared the Solomon Islands strain with Ma07 from Malaysia (5), and several amino acid modifications were detected among samples analyzed in the current study (Table S1). Homologs of this gene have been found in other baculoviruses and nudiviruses, and it is likely to be nonessential, as an insertion/deletion mutation of this gene in BmNPV (Bm98) had no apparent effect on infectivity (25).
OrNV_gp01, which encodes DNA polymerase B, has low expression values in almost all libraries (rank, 97 out of 130 genes), but it is overexpressed (fold change, 2.7; FDR, 0.002) in samples collected from Fiji (Fig. 5C).
OrNV is targeted by the host RNAi response and encodes a putative miRNA.
During virus infection in arthropods, two standard small RNA (sRNA) classes, the microRNAs (miRNAs) and small interfering RNAs (siRNAs), contribute to the antiviral response and promote viral tolerance and, in some cases, clearance of virus infections (reviewed in references 37 and 38). During DNA virus infection, the host riboendonuclease III enzyme Dicer-2 cleaves double-stranded RNA (dsRNA) into virus-derived small interfering RNAs (vsiRNAs) 20 to 22 nt in length (39, 40). These vsiRNAs are loaded into the RNA-induced silencing complex (RISC), where they target RNA molecules through complementarity and reduce DNA virus gene transcription and ultimately virus replication. The RNA interference (RNAi) response in DNA virus infection is subtly different from the RNAi response to RNA virus infections, as dsDNA viruses do not require an RNA genome intermediate for replication. vsiRNAs are abundantly generated from the bidirectional transcription of overlapping gene regions (39) and also secondary RNA structures (41). Some secondary structures of viral RNA are processed by the canonical host miRNA machinery and appear to be functional miRNAs (42). These miRNAs influence virus latency in the case of HzNV (43) but can also autoregulate transcription of DNA virus genes (44).
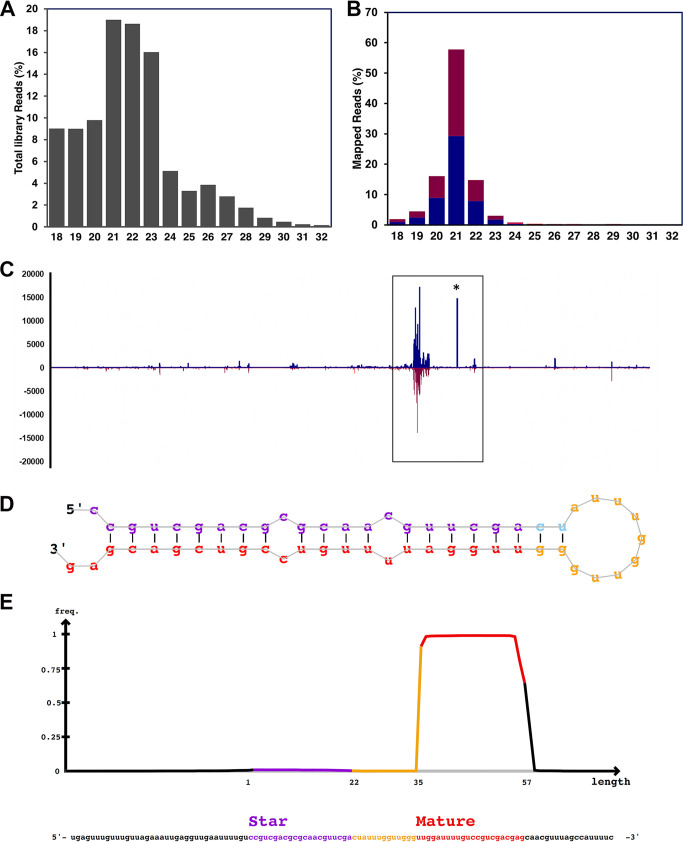
To our knowledge, only one study has examined the virus RNAi response in beetles, with virally derived siRNAs demonstrated in the red flour beetle (Tribolium castaneum) infected with the single-stranded positive-sense RNA Tribolium castaneum iflavirus (Iflaviridae) (45). To examine the repertoire of small RNA responses to OrNV, the small RNA fraction (16 to 32 nt) of a persistently infected sample from the Solomon Islands was subjected to small RNA sequencing and mapped against the OrNV genome. We examined both the size distribution of the virus-derived RNA fragments and “hot spot” genomic locations. After quality and adapter trimming of the small RNA sequencing library (Fig. 6A), 563,774 of the 11,121,239 small RNA (16- to 32-nt) reads (5.07%) could be mapped to both orientations of the OrNV genome. Examination of the size profile of these mapped small RNA reads (Fig. 6B) suggested that the majority (57.7%) correspond to the 21-nt prototypical Dicer-2 vsiRNAs. After examination of the strand origin of the virus-derived small RNA read population, reads originated from both the forward (52.7%) and reverse (47.3%) orientations of the genome more or less evenly. The genomic regions responsible for the high generation of vsiRNAs in the OrNV genome suggested that the biogenesis of vsiRNAs from OrNV was unevenly targeted throughout the OrNV genome, with hot spot regions as previously demonstrated in other DNA-insect RNAi responses (39, 40). This is evidenced by coverage on both forward and reverse sequences similar to a pattern of “mirroring” of vsiRNA between genome positions (Fig. 6C).
FIG 6.
OrNV is targeted by the host RNAi response and encodes a putative miRNA. (A) Length distribution of the small RNA library. (B) Length distribution of reads mapped to the OrNV genome which demonstrated a peak at 21 nt; the red and blue bars represent reads mapped to negative and positive strands. respectively. (C) Noticeable hot spots were detected in several positions on the OrNV genome. The potential miRNA location is shown by an asterisk. (D) OrNV-miR-1 hairpin loop structure. The small RNA read density mapped to different parts of potential OrNV precursor miRNA. (E) A significant number of reads mapped to mature OrNV-miR-1-3p.
One portion of high coverage with vsiRNA reads without a corresponding region of high coverage originated from overlapping negative genome orientation at approximately 85.5 kb of the OrNV genome (Fig. 6C). As a previous report for Drosophila infected with Kallithea virus (Nudiviridae), a close relative of OrNV, suggests that Kallithea virus encodes a miRNA (46), we examined the possibility that OrNV also potentially produces a miRNA. For this small RNA, reads originating from the OrNV genome were analyzed by the miRNA prediction tool mirDeep2 (47). A single putative 22-nt miRNA (UUGGAUUUUGUCCGUCGACGAG) from the 3′ end of a pre-miRNA-like hairpin originating from the genomic location corresponded to a high peak of small RNAs (positions 85464 to 85520 on OrNV_gp098) (Fig. 6C and D). The mature sequence and isomiR variants of this putative miRNA (herein called OrNV-miR-1-3p) were highly abundant in the whole sRNA library and in total, considering 2 nucleotides upstream and 5 nucleotides downstream, amounted to almost 2.6% of all OrNV reads with (14,595 reads).
Additionally, the mature OrNV-miRNA-1 was the single most abundant sRNA sequence of the OrNV-derived small RNAs, with the exact 22 nt accounting for 4,832 reads (0.8%). There were only 65 reads in total associated with the -5p RNA “star” sequence (OrNV-miR-1-5p: CCGUCGACGCGCAACGUUCGA) (Fig. 6E).
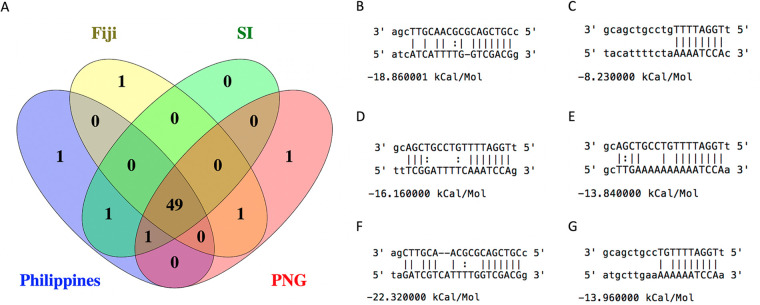
Upon further examination of the miRNA, we found that there was no apparent homology to other coleopteran or arthropod miRNAs as per analysis of similarity to miRNAs annotated on miRBase (release 22.1; October 2018) (48), and the seed region displayed no obvious similarity to known miRNAs. Unfortunately, as the genome of O. rhinoceros is not currently available, we are not able to conduct a preliminary analysis of potential OrNV-miR-1/host gene mRNA interactions; however, miRNA could alternatively regulate virus gene expression of OrNV itself (42). We identified several potential binding sites on the genomes of different geographical strains of OrNV (Fig. 7). Although 49 potential binding sites are universal among all strains, there are some binding sites exclusively identified for Fiji, PNG, and the Philippines which are not detected in other populations (Fig. 7). While numbers of OrNV-miR-1 are far lower than for the 22-nt miRNA reported for Kallithea virus, which represented >35% of all reads (46), the evidence that other large dsDNA viruses with nuclear replication strategies also encode miRNAs and the absence of overlapping transcription or high -5p or “star” reads indicate that OrNV-miR-1 is a promising candidate for future laboratory studies of long-term chronic OrNV persistence in infected O. rhinoceros beetles.
FIG 7.
OrNV-miR-1 binding sites on the genomes of different geographical strains of OrNV. (A) Venn diagram representing the number of common and strain specific binding sites. (B) OrNV-miR-1-3p binding site on gp_027 exclusively in Fiji strains; (C) OrNv-miR-1-5p binding site on gp_069 exclusively in PNG strains; (D) OrNv-miR-1-5p binding site on gp_083 exclusively in Philippines strains; (E) a common OrNv-miR-1-5p binding site on a noncoding part of genome in Fiji and PNG strains; (F) a common OrNv-miR-1-3p binding site on gp_027 in Philippines, Solomon Islands, and PNG strains; (G) a common OrNv-miR-1-5p binding site on a noncoding part of genome in Philippines and Solomon Islands strains.
Conclusion.
We identified a high number of polymorphic sites among several geographical strains of OrNV, but potentially only a few of these variations in the genome are involved in functional changes. We found some genomic variations in OrNV core genes such as OrNV_gp034 (DNA helicase), lef-8, lef-4, and vp91, which can potentially alter their typical function. These findings provide valuable resources for future studies to improve our understanding of the OrNV genetic variations in different geographic regions and their potential link to virus pathogenicity. However, further genomic analyses of OrNV strains in combination with the characterization of its effects on the host across multiple populations are necessary to comprehend if such structural or transcriptional variation is responsible for different levels of effectiveness of OrNV as a biological control agent in different countries.
MATERIALS AND METHODS
Sample collection and processing.
Adult female coconut rhinoceros beetles (O. rhinoceros) were collected from Philippines (Los Baños) and three different South Pacific countries, Fiji (Sigatoka, Viti Levu), Papua New Guinea (Kimbe, New Britain), and Solomon Islands (Honiara, Guadalcanal) using traps baited with aggregation pheromone (Oryctalure, P046-Lure; ChemTica Internacional, S. A., Heredia, Costa Rica) between June and October 2019.
Individual female insects were disinfected by soaking in 75% ethanol for 30s and then rinsing in phosphate-buffered saline (PBS) before their gut tissues were dissected, removed, and preserved in an RNA stabilization reagent (RNAlater; Qiagen). For each individual, some fat body and small pieces of gut tissues were also preserved separately in 95% ethanol. Preserved specimens were shipped to the University of Queensland, Brisbane, Australia, for further analysis. The preserved gut tissues were kept at −80°C upon arrival. For examination of virus infection status and mitochondrial haplotype, DNA was extracted from fat bodies and gut fragments using the Qiagen blood and tissue DNA extraction kit according to the manufacturer’s instructions.
The presence of the OrNV was confirmed by successful amplification of a 945-bp product using the OrV15 primers that target the OrNV_gp083 gene (49) and by Sanger sequencing of each PCR product. For confirmation of the mitochondrial haplotype, a small fragment of the CoxI gene was amplified in these individuals with the primers LCO1490 and HCO2198 (50). PCR products were sequenced bidirectionally using an ABI3730 genetic analyzer (Applied Biosystems) at Macrogen Inc., Seoul, South Korea.
The guts from OrNV-positive individuals were further dissected and transferred to Qiazol lysis reagent for RNA extraction according to the manufacturer’s instructions (Qiagen; catalog no. 79306). The RNA samples were treated with DNase I for 1 h at 37°C, and then their concentrations were measured using a spectrophotometer and integrity was ensured through analysis of RNA on a 1% (wt/vol) agarose gel. After the RNA quality was checked, total RNA samples were submitted to Novogene Genomics Singapore Pte Ltd. for RNA sequencing (RNA-seq). The PCR-based cDNA libraries were prepared using the NEBNext Ultra RNA library prep kit and were sequenced using Novaseq6000 (PE150) technology with an insert size between 250 and 300 bp.
Bioinformatic analysis.
The CLC Genomics Workbench version 12.0.1 was used for bioinformatic analyses. All libraries were trimmed from any vector or adapter sequences remaining. Low-quality reads (quality score below 0.05) and reads with more than two ambiguous nucleotides were discarded. For identification of the virus consensus from each sample, clean reads mapped to a recently reported Oryctes rhinoceros nudivirus complete genome sequence (MN623374) (5). We implemented strict mapping criteria (mismatch, insertion, and deletion costs, 2, 3, and 3, respectively). A minimum similarity and length fraction of 0.9 between a mapped segment and the reference was allowed in mapping criteria. We also permitted a minimum of 5 reads as a low coverage threshold and inserted N as ambiguity symbols for handling the low-coverage region. The quality score for each nucleotide has been used to resolve any conflict. For multiple-sequence alignments, we used MUltiple Sequence Comparison by Log-Expectation (MUSCLE v3.8.425) to compare OrNV whole-genome consensus sequence from different libraries.
Genomic variant and phylogenetic analysis.
To detect genomic modifications among different OrNV-infected O. rhinoceros populations, the CLC Genomic Workbench low-frequency variant detection tool, which is based on a neighborhood quality standard (NQS) algorithm, was used. Variants such as single nucleotide variants (SNV), multiple nucleotide variants (MNV), insertion, deletion, and amino acid modification were then called based on neighborhood radius (5), a maximum number of gaps and mismatches (2), the minimum average quality of surrounding bases (15), and minimum quality of the central base (20). We also used a minimum coverage filter of 10 and a minimum variant frequency of 35% to exclude sequencing artifacts. The final alignments for downstream analysis were OrNV 11 × 126,183 nt and for the partial CoxI 11 × 621 nt. For the construction of the maximum likelihood phylogenetic tree of both host mitochondrial DNA and OrNV, we selected a nucleotide substitution model based on the consensus outcome of hierarchical likelihood ratio test (hLRT), Bayesian information criterion (BIC), and Akaike information criterion (AIC). For the OrNV phylogenetic inference, the general time reversible (GTR) +G rate variation (4 categories), +T topology variation was selected from all tests. For the mitochondrial CoxI, the Hasegawa-Kishino-Yano (HKY) substitution model with +T topology variation was selected. The maximum likelihood phylogeny was then constructed using CLC Genomics Workbench with a neighbor-joining starting tree construction method under the nucleotide substitution models mentioned above with 1,000 bootstraps. Resultant trees were exported and visualized using FigTree version 1.4 (A. Rambaut; http://tree.bio.ed.ac.uk/software/figtree/). Tests for recombination between multiple-sequence alignments of the individual 139 genes of OrNV were conducted using the RDP4 program (51) using the automated RDP, GENECOV (52), and Bootscan (51) algorithms.
Viral gene expression profiling.
For examination of OrNV gene expression profiles in different O. rhinoceros populations, clean reads were mapped to the OrNV genome using the RNA Sequencing Tool on CLC genomics workbench. Expression values for all OrNV genes were calculated as total mapped read count and then normalized to reads per kilobase of transcript per million mapped viral reads (RPKM). The most highly expressed OrNV genes in O. rhinoceros gut tissue in different populations were identified and ranked. The viral gene expression profile was created based on calculated RPKM values. For comparison of viral gene expression among diverse OrNV-infected O. rhinoceros populations, hierarchical clustering analysis with Euclidean distance metric on the log2-transformed RPKM value (normalized expression value) was performed by CLC Genomic Workbench. We considered genes with >2-fold changes and a false-discovery rate (FDR) of less than 0.05 as significantly modulated viral genes. For further functional annotation of OrNV hypothetical proteins, we searched for a conserved domain through IntrProScan database (53). We also used EggNOG version 4.5 to run pairwise orthology prediction in Cluster of Orthologous Groups (54).
Analysis of virus-derived small RNA and OrNV miRNA prediction.
For analysis of the host RNAi response to OrNV, a small RNA library was generated from a virus-infected individual from the Solomon Islands using the NEBNext multiplex small RNA library prep kit for Illumina at the Novogene Genomics Singapore Pte Ltd. The purified cDNA libraries were sequenced on Novaseq 6000 (SE50), and raw sequencing reads were obtained using Illumina’s Sequencing Control Studio software. Raw data were stripped of adapters, and reads with a quality score above 0.05 and less than two ambiguous nucleotides were retained. Reads without 3′ adapters and also reads with less than 16 nt were discarded. The clean reads mapped to the recently sequenced OrNV genome (MN623374). For examination of potential virus-derived miRNAs of OrNV, we used miRDeep v2.0.0.8 (47) hosted on the Mississippi Galaxy instance. We screened the genome consensus of all OrNV geographical strains for potential miRNA binding sites with miRanda algorithm (55). This tool considers matching along the entire miRNA sequence, but we ran the program in strict mode, which demands strict 5′ seed pairing.
Data availability.
Deep sequencing raw data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE152658.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by Australian Centre for International Agricultural Research funding (HORT/2016/185) and the University of Queensland (UQECR2057321).
We thank Apenisa Sailo (Ministry of Agriculture, Fiji), Helen Tsatsia (Ministry of Agriculture and Livestock, Solomon Islands), and colleagues from the Papua New Guinea Oil Palm Research Association, Dami research station, for their assistance in collecting and providing the insect specimens.
Kayvan Etebari, Conceptualization, Methodology, Investigation, Formal Analysis, Data Curation, Visualization, Writing—Original Draft Preparation, and Funding Acquisition; Rhys Parry, Data Curation, Formal Analysis, Visualization, and Writing—Original Draft Preparation; Marie Joy B. Beltran, Resources; Michael J. Furlong, Conceptualization, Writing—Reviewing and Editing, Funding Acquisition, Project Administration, and Supervision.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Harrison RL, Herniou EA, Bezier A, Jehle JA, Burand JP, Theilmanns DA, Krell PJ, van Oers MM, Nakai M, ICTV Report Consortium. 2020. ICTV virus taxonomy profile: Nudiviridae. J Gen Virol 101:3–4. doi: 10.1099/jgv.0.001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jehle JA. 2010. Nudiviruses: their biology and genetics, p 153–168. In Asgari S, Johnson KN (ed), Insect virology. Caister Academic Press, London, United Kingdom. [Google Scholar]
- 3.Yang Y-T, Lee D-Y, Wang Y, Hu J-M, Li W-H, Leu J-H, Chang G-D, Ke H-M, Kang S-T, Lin S-S, Kou G-H, Lo C-F. 2014. The genome and occlusion bodies of marine Penaeus monodon nudivirus (PmNV, also known as MBV and PemoNPV) suggest that it should be assigned to a new nudivirus genus that is distinct from the terrestrial nudiviruses. BMC Genomics 15:e628. doi: 10.1186/1471-2164-15-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang YJ, Kleespies RG, Ramle MB, Jehle JA. 2008. Sequencing of the large dsDNA genome of Oryctes rhinoceros nudivirus using multiple displacement amplification of nanogram amounts of virus DNA. J Virol Methods 152:106–108. doi: 10.1016/j.jviromet.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Etebari K, Filipovic I, Rasic G, Devine GJ, Tsatsia H, Furlong MJ. 2020. Complete genome sequence of Oryctes rhinoceros nudivirus isolated from the coconut rhinoceros beetle in Solomon Islands. Virus Res 278:197864. doi: 10.1016/j.virusres.2020.197864. [DOI] [PubMed] [Google Scholar]
- 6.Bezier A, Theze J, Gavory F, Gaillard J, Poulain J, Drezen J-M, Herniou EA. 2015. The genome of the nucleopolyhedrosis-causing virus from Tipula oleracea sheds new light on the Nudiviridae family. J Virol 89:3008–3025. doi: 10.1128/JVI.02884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedford GO. 2013. Biology and management of palm dynastid beetles: recent advances. Annu Rev Entomol 58:353–372. doi: 10.1146/annurev-ento-120710-100547. [DOI] [PubMed] [Google Scholar]
- 8.Huger AM. 1966. A virus disease of the Indian rhinoceros beetle, Oryctes rhinoceros (Linnaeus), caused by a new type of insect virus, Rhabdionvirus oryctes. J Invertebr Pathol 8:38–51. doi: 10.1016/0022-2011(66)90101-7. [DOI] [PubMed] [Google Scholar]
- 9.Etebari K, Hereward J, Sailo A, Ahoafi EM, Tautua R, Tsatsia H, Jackson GV, Furlong MJ. 2020. Genetic structure of the coconut rhinoceros beetle (Oryctes rhinoceros) population and the incidence of its biocontrol agent (Oryctes rhinoceros nudivirus) in the South Pacific Islands. bioRxiv doi: 10.1101/2020.07.30.229872. [DOI] [PMC free article] [PubMed]
- 10.Marshall SDG, Moore A, Vaqalo M, Noble A, Jackson TA. 2017. A new haplotype of the coconut rhinoceros beetle, Oryctes rhinoceros, has escaped biological control by Oryctes rhinoceros nudivirus and is invading Pacific Islands. J Invertebr Pathol 149:127–134. doi: 10.1016/j.jip.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Zelazny B, Alfiler AR, Lolong A. 1989. Possibility of resistance to a baculovirus in populations of the coconut rhinoceros beetle (Oryctes rhinoceros). FAO Plant Prot Bull 37:77–82. [Google Scholar]
- 12.Zelazny B, Alfiler AR. 1991. Ecology of baculovirus‐infected and healthy adults of Oryctes rhinoceros (Coleoptera: Scarabaeidae) on coconut palms in the Philippines. Ecol Entomol 16:253–259. doi: 10.1111/j.1365-2311.1991.tb00215.x. [DOI] [Google Scholar]
- 13.Zelazny B, Lolong A, Pattang B. 1992. Oryctes rhinoceros (Coleoptera: Scarabaeidae) populations suppressed by a baculovirus. J Invertebr Pathol 59:61–68. doi: 10.1016/0022-2011(92)90112-H. [DOI] [Google Scholar]
- 14.Crawford AM, Zelazny B. 1990. Evolution in Oryctes baculovirus: rate and types of genomic change. Virology 174:294–298. doi: 10.1016/0042-6822(90)90078-6. [DOI] [PubMed] [Google Scholar]
- 15.Moslim R, Kamarudin N, Ghani IA, Wahid MB, Jackson TA, Tey CC, Ahdly M. 2011. Molecular approaches in the assessment of Oryctes rhinoceros virus for the control of rhinoceros beetle in oil palm plantations. J Oil Palm Res 23:1096–1109. [Google Scholar]
- 16.Ramle M, Wahid MB, Norman K, Glare TR, Jackson TA. 2005. The incidence and use of Oryctes virus for control of rhinoceros beetle in oil palm plantations in Malaysia. J Invertebr Pathol 89:85–90. doi: 10.1016/j.jip.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Jackson TA, Crawford AM, Glare TR. 2005. Oryctes virus—time for a new look at a useful biocontrol agent. J Invertebr Pathol 89:91–94. doi: 10.1016/j.jip.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Loiseau V, Herniou EA, Moreau Y, Leveque N, Meignin C, Daeffler L, Federici B, Cordaux R, Gilbert C. 2020. Wide spectrum and high frequency of genomic structural variation, including transposable elements, in large double-stranded DNA viruses. Virus Evolution 6:vez060. doi: 10.1093/ve/vez060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chateigner A, Bezier A, Labrousse C, Jiolle D, Barbe V, Herniou EA. 2015. Ultra deep sequencing of a baculovirus population reveals widespread genomic variations. Viruses 7:3625–3646. doi: 10.3390/v7072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theze J, Cabodevilla O, Palma L, Williams T, Caballero P, Herniou EA. 2014. Genomic diversity in European Spodoptera exigua multiple nucleopolyhedrovirus isolates. J Gen Virol 95:2297–2309. doi: 10.1099/vir.0.064766-0. [DOI] [PubMed] [Google Scholar]
- 21.Shrestha A, Bao K, Chen Y-R, Chen W, Wang P, Fei Z, Blissard GW. 2018. Global analysis of baculovirus Autographa californica multiple nucleopolyhedrovirus gene expression in the midgut of the lepidopteran host Trichoplusia ni. J Virol 92:e01277-18. doi: 10.1128/JVI.01277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cory JS, Green BM, Paul RK, Hunter-Fujita F. 2005. Genotypic and phenotypic diversity of a baculovirus population within an individual insect host. J Invertebr Pathol 89:101–111. doi: 10.1016/j.jip.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Hill T, Unckless RL. 2018. The dynamic evolution of Drosophila innubila nudivirus. Infect Genet Evol 57:151–157. doi: 10.1016/j.meegid.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill T, Unckless RL. 2017. Baculovirus molecular evolution via gene turnover and recurrent positive selection of key genes. J Virol 91:e01319-17. doi: 10.1128/JVI.01319-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ono C, Kamagata T, Taka H, Sahara K, Asano S, Bando H. 2012. Phenotypic grouping of 141 BmNPVs lacking viral gene sequences. Virus Res 165:197–206. doi: 10.1016/j.virusres.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Ioannidis K, Swevers L, Iatrou K. 2016. Bombyx mori nucleopolyhedrovirus lef8 gene: effects of deletion and implications for gene transduction applications. J Gen Virol 97:786–796. doi: 10.1099/jgv.0.000383. [DOI] [PubMed] [Google Scholar]
- 27.Hitchman RB, Possee RD, King LA. 2012. High-throughput baculovirus expression in insect cells. Methods Mol Biol 824:609–627. doi: 10.1007/978-1-61779-433-9_33. [DOI] [PubMed] [Google Scholar]
- 28.Cheng RL, Xi Y, Lou YH, Wang Z, Xu JY, Xu HJ, Zhang CX. 2014. Brown planthopper nudivirus DNA integrated in its host genome. J Virol 88:5310–5318. doi: 10.1128/JVI.03166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burke GR. 2019. Common themes in three independently derived endogenous nudivirus elements in parasitoid wasps. Curr Opin Insect Sci 32:28–35. doi: 10.1016/j.cois.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, Li C-X, Qin X-C, Li J, Cao J-P, Eden J-S, Buchmann J, Wang W, Xu J, Holmes EC, Zhang Y-Z. 2016. Redefining the invertebrate RNA virosphere. Nature 540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 31.Bazan JF, Fletterick RJ. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci U S A 85:7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babe LM, Craik CS. 1997. Viral proteases: evolution of diverse structural motifs to optimize function. Cell 91:427–430. doi: 10.1016/s0092-8674(00)80426-2. [DOI] [PubMed] [Google Scholar]
- 33.Palmer WH, Joosten J, Overheul GJ, Jansen PW, Vermeulen M, Obbard DJ, Van Rij RP. 2019. Induction and suppression of NF-κB signalling by a DNA virus of Drosophila. J Virol 93:e01443-18. doi: 10.1128/JVI.01443-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou D, Kuang W, Luo S, Zhang F, Zhou F, Chen T, Zhang Y, Wang H, Hu Z, Deng F, Wang M. 2019. Baculovirus ODV-E66 degrades larval peritrophic membrane to facilitate baculovirus oral infection. Virology 537:157–164. doi: 10.1016/j.virol.2019.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Xiang XW, Chen L, Hu XL, Yu SF, Yang R, Wu XF. 2011. Autographa californica multiple nucleopolyhedrovirus odv-e66 is an essential gene required for oral infectivity. Virus Res 158:72–78. doi: 10.1016/j.virusres.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Simon O, Williams T, Cerutti M, Caballero P, Lopez-Ferber M. 2013. Expression of a peroral infection factor determines pathogenicity and population structure in an insect virus. PLoS One 8:e0078834. doi: 10.1371/journal.pone.0078834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asgari S. 2018. microRNAs as regulators of insect host-pathogen interactions and immunity. Adv Insect Physiol 55:19–45. doi: 10.1016/bs.aiip.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Bronkhorst AW, van Rij RP. 2014. The long and short of antiviral defense: small RNA-based immunity in insects. Curr Opin Virol 7:19–28. doi: 10.1016/j.coviro.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Jayachandran B, Hussain M, Asgari S. 2012. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J Virol 86:13729–13734. doi: 10.1128/JVI.02041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabin LR, Zheng Q, Thekkat P, Yang J, Hannon GJ, Gregory BD, Tudor M, Cherry S. 2013. Dicer-2 processes diverse viral RNA species. PLoS One 8:e55458. doi: 10.1371/journal.pone.0055458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parry R, Bishop C, De Hayr L, Asgari S. 2019. Density-dependent enhanced replication of a densovirus in Wolbachia-infected Aedes cells is associated with production of piRNAs and higher virus-derived siRNAs. Virology 528:89–100. doi: 10.1016/j.virol.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Hussain M, Asgari S. 2010. Functional analysis of a cellular microRNA in insect host-ascovirus interaction. J Virol 84:612–620. doi: 10.1128/JVI.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu YL, Wu CP, Liu CYY, Hsu PWC, Wu EC, Chao YC. 2011. A non-coding RNA of insect HzNV-1 virus establishes latent viral infection through microRNA. Sci Rep 1:e00060. doi: 10.1038/srep00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain M, Taft RJ, Asgari S. 2008. An insect virus-encoded microRNA regulates viral replication. J Virol 82:9164–9170. doi: 10.1128/JVI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, Smith SA, Sharma PP, Cordaux R, Gilbert C, Giraud I, Collins DH, Zamore PD, Miska EA, Sarkies P, Jiggins FM. 2018. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol 2:174–181. doi: 10.1038/s41559-017-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster CL, Waldron FM, Robertson S, Crowson D, Ferrari G, Quintana JF, Brouqui J-M, Bayne EH, Longdon B, Buck AH, Lazzaro BP, Akorli J, Haddrill PR, Obbard DJ. 2015. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol 13:e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozomara A, Birgaoanu M, Griffiths-Jones S. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richards NK, Glare TR, Aloalií I, Jackson TA. 1999. Primers for the detection of Oryctes virus from Scarabaeidae (Coleoptera). Mol Ecol 8:1552–1553. doi: 10.1046/j.1365-294x.1999.07072.x. [DOI] [PubMed] [Google Scholar]
- 50.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299. [PubMed] [Google Scholar]
- 51.Martin DP, Posada D, Crandall KA, Williamson C. 2005. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. Aids Res Hum Retroviruses 21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- 52.Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- 53.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang H-Y, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong S-Y, Finn RD. 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol 5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep sequencing raw data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE152658.