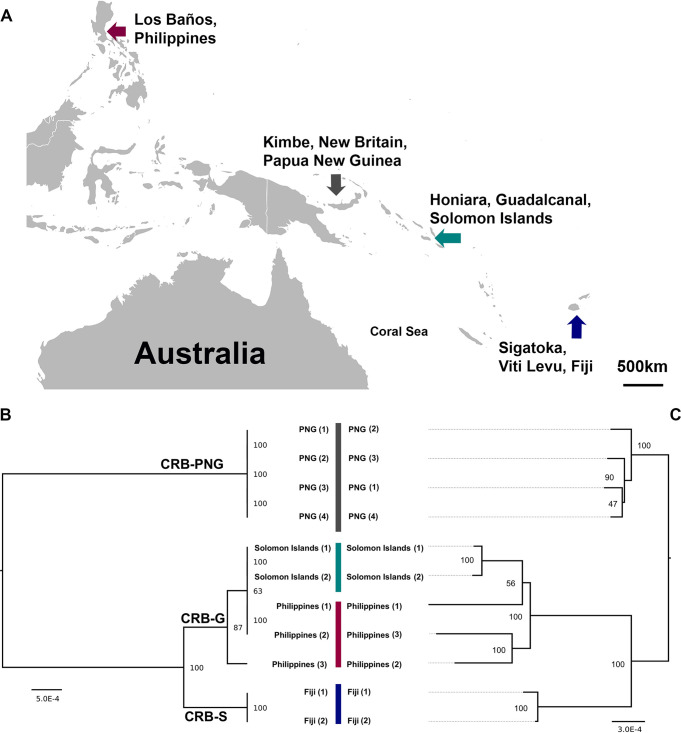
FIG 1.
(A) Geographic locations of coconut rhinoceros beetles (O. rhinoceros) chronically infected with OrNV from Southeast Asia and the Pacific Islands. (B) Host mitochondrial maximum likelihood phylogeny based on O. rhinoceros CoxI gene sequence. Four different haplotypes of O. rhinoceros in three major mitochondrial lineages (CRB-PNG, CRB-S and CRB-G) were identified. (C) Maximum likelihood phylogeny analysis of OrNV strains. Samples from the Solomon Islands and Philippines are closely related to each other, while individuals from PNG and Fiji are separated from them and located in another clade. Harmonious topologies of both host and OrNV strains provide evidence of codivergence of OrNV strains with their O. rhinoceros hosts. Branch length indicates nucleotide substitutions per site; trees were bootstrapped 1,000 times. The host phylogenetic tree is midpoint rooted.