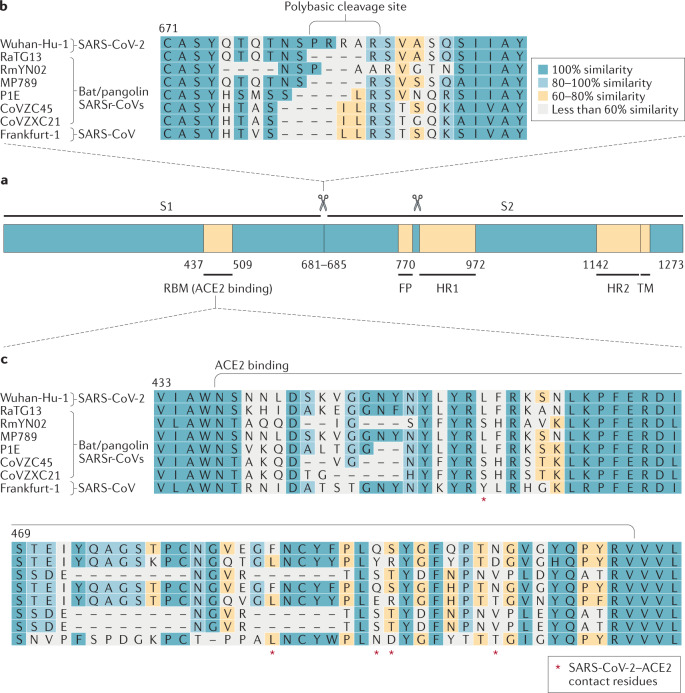
Fig. 2. Severe acute respiratory syndrome-related coronavirus spike sequence variation.
a | Schematic illustration of coronavirus spike, indicating domain 1 and domain 2. The receptor-binding motif (RBM) is located on S1 and the fusion peptide (FP), heptad repeat 1 (HR1), HR2 and the transmembrane (TM) domains are located on S2. The cleavage sites are indicated. The colour code designates conserved spike regions surrounding the angiotensin-converting enzyme 2 (ACE2)-binding domain among severe acute respiratory syndrome-related coronaviruses (SARSr-CoVs) and high amino acid sequence variations within the site of receptor interaction. b | Amino acid alignment of human SARS-CoV-2 (Wuhan-Hu-1) and SARS-CoV (Frankfurt-1), bat (RaTG13, RmYN02, CoVZC45 and CoVZXC21) and pangolin (MP789, P1E) SARSr-CoVs. The spike gene sequence alignment was performed using MUSCLE and using the default settings and codon alignment, then translated into amino acids using MEGA7, version 7.0.26. The alignment was coloured according to percentage amino acid similarity with a Blosum 62 score matrix. The colour code designates conserved spike regions surrounding the ACE2-binding domain among SARSr-CoVs and high amino acid sequence variations within the site of receptor interaction. The insertion of a polybasic cleavage site (PRRAR, amino acids 681 to 685) in Wuhan-Hu-1 is indicated, and similar insertions are depicted in bat SARSr-CoV RmYN02. c | Within the spike sequence, the ACE2 receptor-binding motif (amino acids 437 to 509, black line) is depicted. The spike contact residues for ACE2 interaction are marked with asterisks.