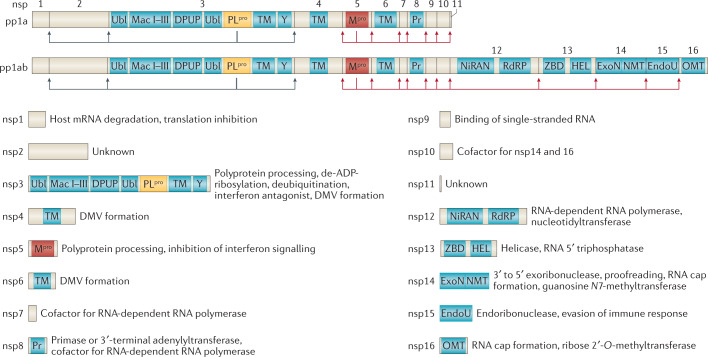
Fig. 3. Coronavirus polyprotein processing and non-structural proteins.
Coronavirus polyprotein processing and domains of non-structural proteins (nsp) are illustrated for severe acute respiratory syndrome-related coronaviruses. Proteolytic cleavage of the polyproteins pp1a and pp1ab is facilitated by viral proteases residing in nsp3 (PLpro) and nsp5 (Mpro). PLpro proteolytically releases nsp1, nsp2, nsp3 and the amino terminus of nsp4 from the polyproteins pp1a and pp1ab (indicated by the blue arrows). Mpro proteolytically releases nsp5–16 and the carboxy terminus of nsp4 from the polyproteins pp1a and pp1ab (indicated by the red arrows)176. Conserved domains and known functions are schematically depicted for nsp1–16 (refs4,66,67,177). DMV, double-membrane vesicle; DPUP, Domain Preceding Ubl2 and PLpro; EndoU, endoribonuclease; ExoN, exoribonuclease; HEL, helicase; Mac I–III, macrodomains 1–3; Mpro, main protease; NiRAN, nidovirus RdRP-associated nucleotidyltransferase; NMT, guanosine N7-methyltransferase; OMT, ribose 2′-O-methyltransferase; PLpro, papain-like protease; Pr, primase or 3′-terminal adenylyl-transferase; RdRP, RNA-dependent RNA polymerase; TM, transmembrane domains; Ubl, ubiquitin-like domain; Y, Y and CoV-Y domain; ZBD, zinc-binding domain.