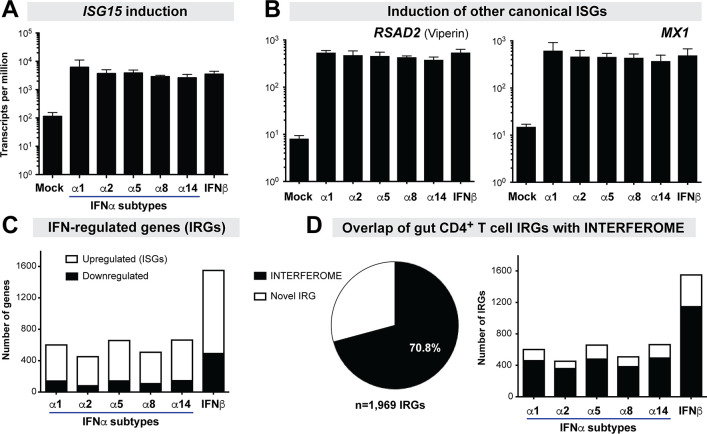
Fig 3. Identification of Novel IRGs.
LPMCs (n = 3 donors) were stimulated with IFNα subtypes and IFNβ normalized to ISG15 ISRE-activity and the transcriptomes were evaluated at 18 h via RNAseq. Sequences were compared against annotated Ensembl genes (version GRCh38). IRGs were defined based on a 1.5-fold change and an FDR cut-off of 20% relative to mock. Induction of (A) ISG15 and (B) RSAD2 and MX1 by IFN-Is based on the RNAseq data. (C) Number of upregulated versus downregulated IRGs. (D) Overlap of gut CD4+ T cell ‘interferomes’ with published IRGs in the INTERFEROME database 2.0 [5].