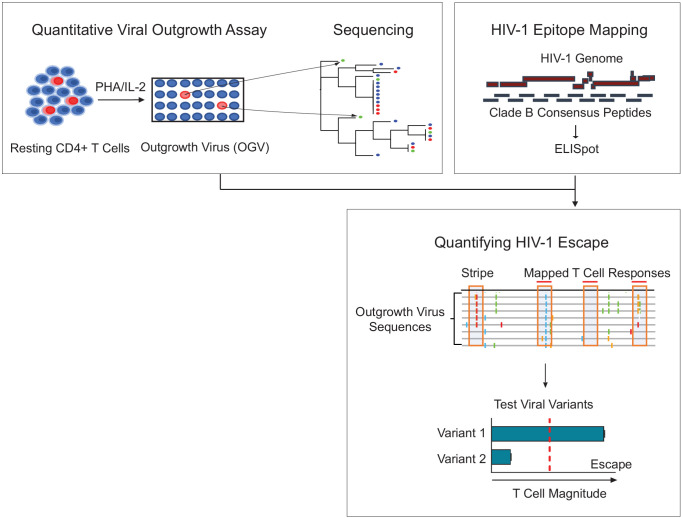
Figure 1. Schema of empirical testing of T cell escape in the HIV-1 reservoir.
Top left panel. The quantitative viral outgrowth assay (QVOA) provided a minimal estimate of the size of the replication-competent reservoir virus from total resting CD4+ T cells of PLWH on ART (n = 25). Supernatants from HIV-1 p24+ wells harboring outgrowth virus (OGV) were sequenced by either PacBio (n = 22) or MiSeq (n = 1). Top right panel. Ex vivo IFN-γ ELISpot assays identified reactive T cell epitopes in clade B HIV-1 proteome in each participant. Lower panel. Sequencing and T cell data were combined to identify putative virus escape mutations by overlaying reactive T cell reactive epitopes (red horizontal line) on highlighter plots of each participant’s outgrowth sequences (n = 23). Peptides spanning all non-synonymous changes between the clade B-defined epitope and outgrowth sequences (orange boxes) were synthesized. In addition, peptides matching OGVs in participants in which > 40% of viruses contained the same non-synonymous mutation generating a ‘stripe’ on the highlighter plot were also synthesized (orange box, no overhead red line). Variant peptides were examined for their impact on T cell recognition in ex vivo IFN-γ ELISpot. Virus escape was defined as a ≥ 50% difference in magnitude between variants, either Clade B sequence or OGV variants (vertical, red dotted line).