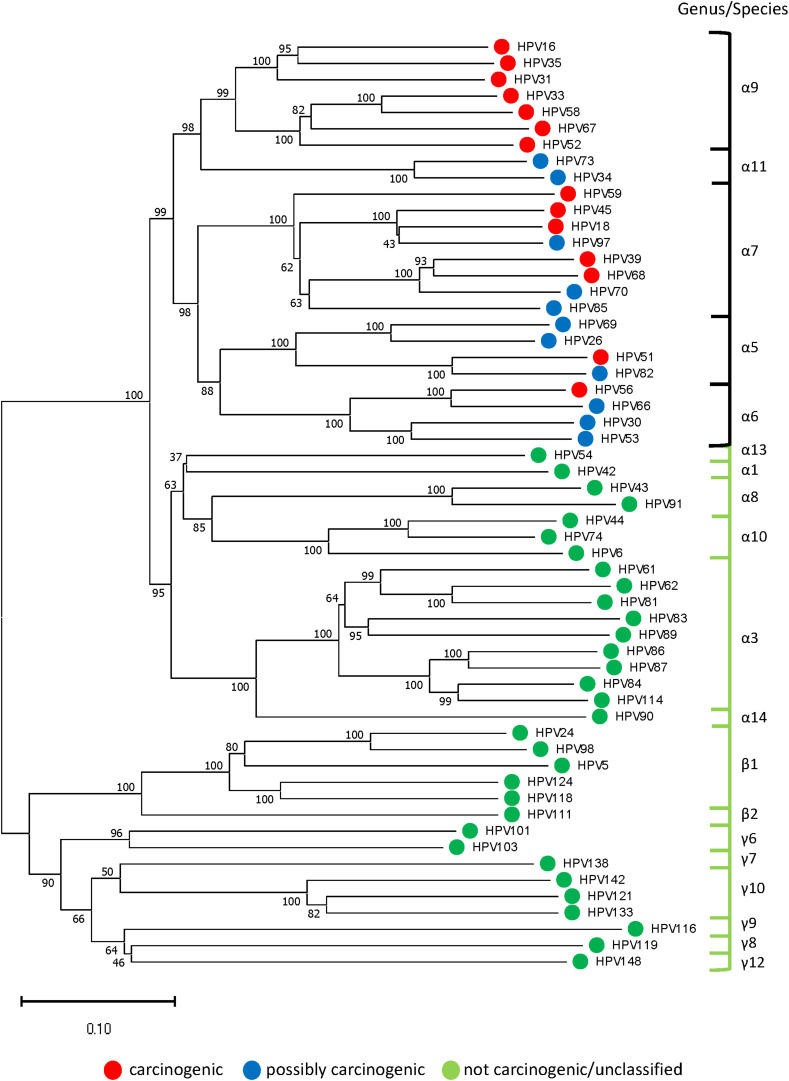
FIGURE 3.
Representative phylogenetic tree of HPV genotypes identified in the clinical samples. Neighbor-Joining tree of 57 HPV genotypes (one from each genotype) revealed two distinct clades in the alpha genera: “high-risk” containing carcinogenic and possibly carcinogenic types [black bracket] and “low-risk” containing probably not carcinogenic or not classifiable types [green bracket]. The beta and gamma genera formed another clade composed of commensal and unclassified genotypes. With HPV-16 at the pinnacle of HPV carcinogenic potential, genetic divergence from this point correlated with decreased carcinogenic risk (phenotype) and grade of cytopathology. The evolutionary history was inferred using the Neighbor-Joining method after concatenating 57 aligned, E6, E7, and L1 reference nucleotide sequences from Papillomavirus Episteme by MUSCLE (Edgar, 2004). The optimal tree with the sum of branch length = 9.99536245 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 2850 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018).