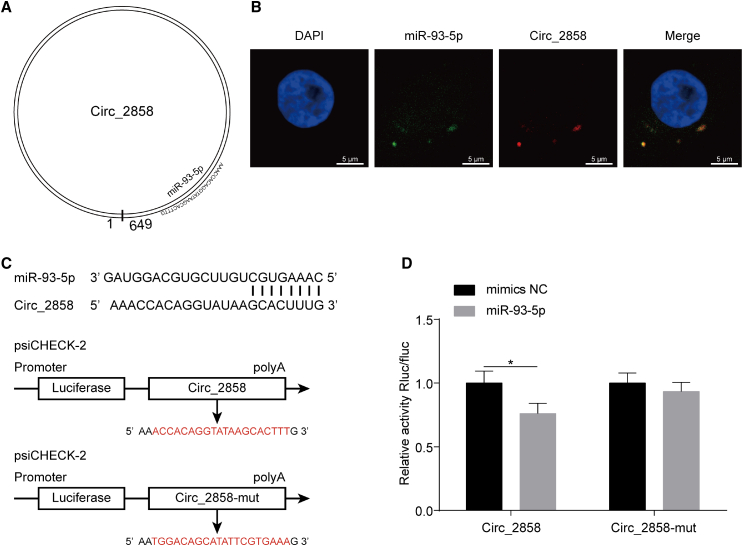
Figure 5.
circ_2858 Sponges with miR-93-5p
(A) Schematic drawing showing the putative binding sites of miR-93-5p on circ_2858. (B) Co-localization of miR-93-5p and circ_2858 in hBMECs by RNA FISH assay. Nuclei were stained with DAPI. miR-93-5p was probed with FAM (green), while circ_2858 was probed with Cy3 (red). Scale bars, 5 μm. (C) Putative miR-93-5p-binding site on circ_2858. These binding sequences and the respective mutant sequences (labeled in red) were cloned into the psiCHECK-2 vector. (D) Dual-luciferase reporter assays showing that miR-93-5p significantly reduced the luciferase activity of the wild-type circ_2858 construct but not of the mutant construct. The Renilla luciferase activity was normalized to the firefly luciferase activity, and data are presented as mean ± SD from three independent experiments.